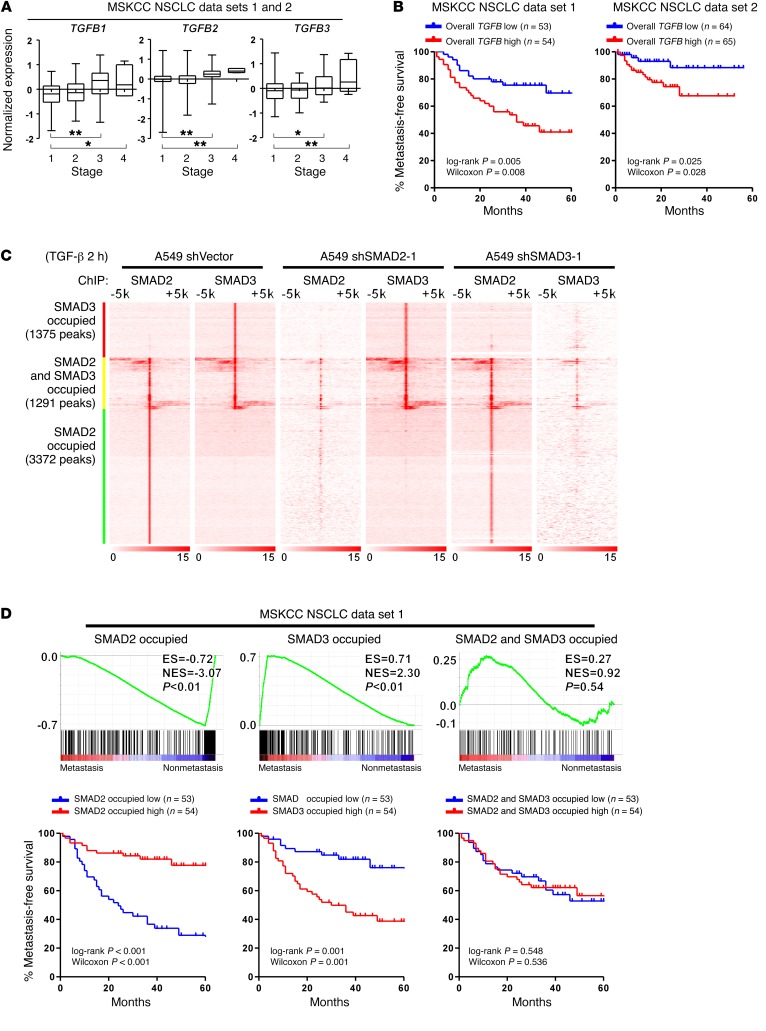
Figure 1. NSCLC metastasis is associated with inactivation of SMAD2-mediated and activation of SMAD3-mediated transcriptional programs.
(A) Analysis of publicly available MSKCC NSCLC data sets 1 and 2 indicates that the mRNA expression of TGFB1, TGFB2, and TGFB3 is significantly increased in patients with stage 3 or stage 4 NSCLC. n = 153 cases (stage 1); n = 35 cases (stage 2); n = 43 cases (stage 3); and n = 5 cases (stage 4). Box and whiskers plots represent the mRNA levels. *P < 0.05 and **P < 0.01, by 1-way ANOVA. (B) Kaplan-Meier analysis indicates that when stratifying patients by the median level of overall TGFB (average of TGFB1, TGFB2, and TGFB3), a high level of overall TGFB (average of TGFB1, TGFB2, and TGFB3) is associated with a shorter metastasis-free survival time in patients in the MSKCC NSCLC data sets 1 and 2. (C) Heatmap of the ChIP enrichment signal surrounding SMAD2 and SMAD3 peaks shows the chromosomal distribution of SMAD2 and SMAD3 in the indicated A549 cells. (D) GSEA and Kaplan-Meier analysis indicate that attenuated SMAD2-specific and enhanced SMAD3-specific transcripts are present in NSCLC patients with an increased metastatic potential in association with a shorter metastasis-free survival in the MSKCC NSCLC data set 1, whereas targeted genes shared by both SMAD2 and SMAD3 are not associated with metastasis status. The definition of “high” and “low” expression of each gene set was stratified by the median of the normalized expression levels of each gene in the set. ES, enrichment score; NES, normalized enrichment score.