Figure I. Schematic of the steps in ribosome profiling.
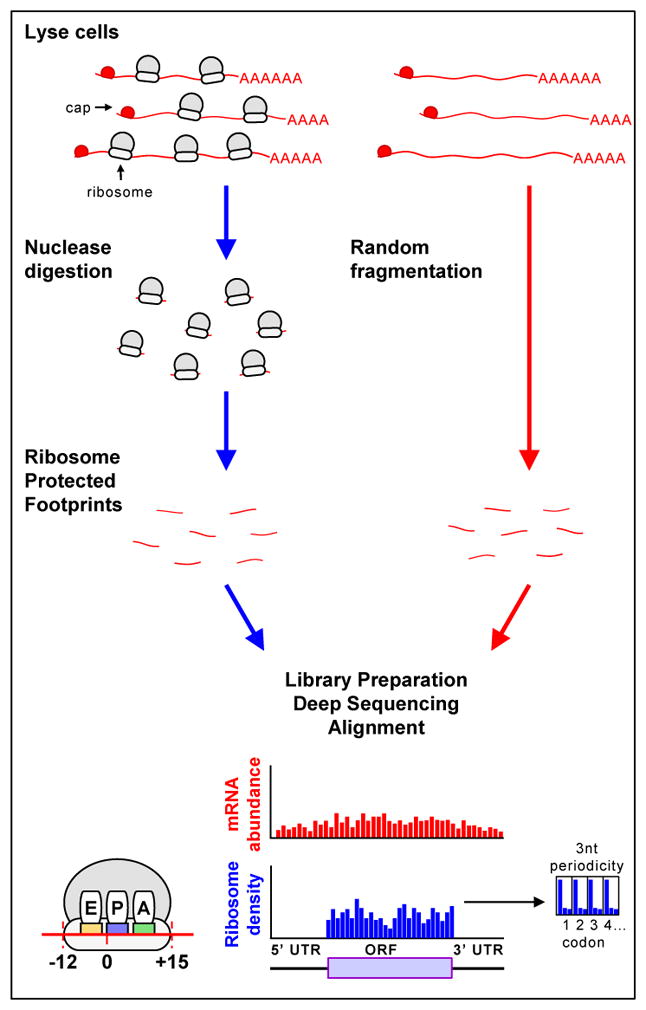
To obtain ribosome footprints, cell lysates, obtained from cells in which translation has been rapidly stopped, are treated with nucleases to digest unprotected RNAs. Ribosome-protected fragments of discrete sizes (22–28 nucleotides in eukaryotes and 15–45 nucleotides in bacteria) are isolated, converted to a library and subjected to deep sequencing. To obtain information on the identity and amounts of mRNA, mRNA is subjected to random fragmentation, isolated, converted to a similar library and sequenced. Sequences are aligned to the genome, such that codons in the A, P and E sites of the ribosome are distinguished. As expected, most ribosome footprints (blue) map to sequences that encode proteins (beginning with the AUG initiation codon and ending at the stop codon) and exhibit a 3 nucleotide periodicity (shown to right of ribosome reads); mRNA footprints (red) extend outside of the translated regions and do not exhibit this periodicity. UTR: untranslated region; ORF: open reading frame.
