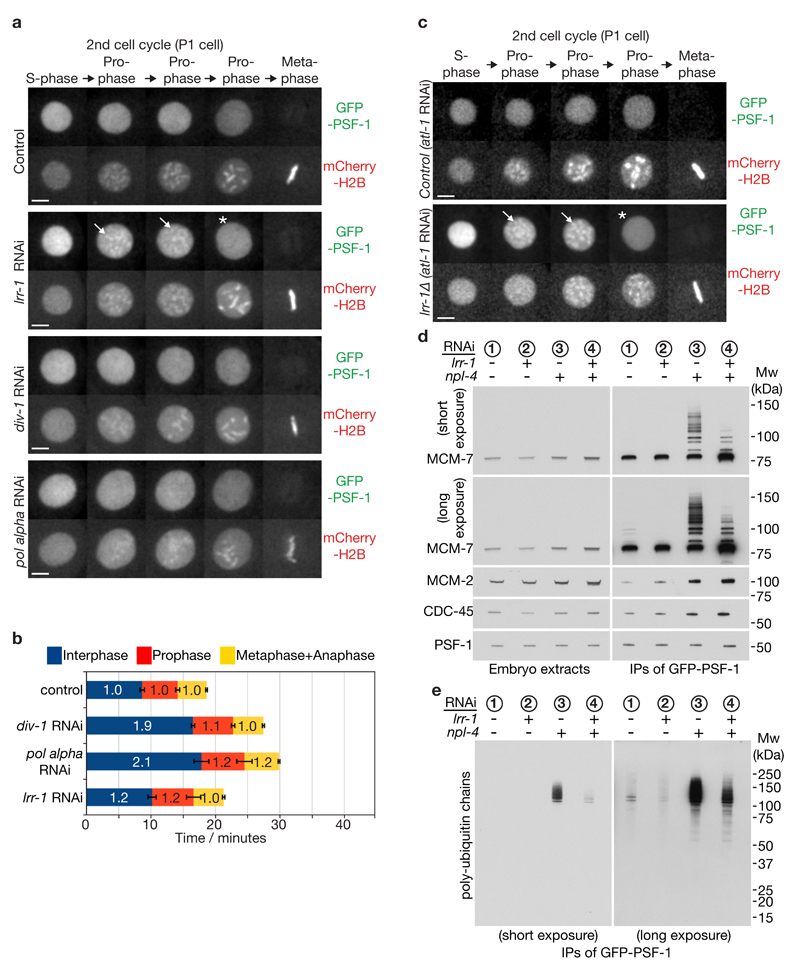
Figure 3. A mitotic pathway for CMG helicase disassembly is revealed in the absence of CUL-2LRR-1.
(a) Embryos from GFP-psf-1 mCherry-H2B worms were exposed to the indicated RNAi treatments, or empty vector in the control, and then processed as in Figure 1b, except that the figure depicts data from the second embryonic cell cycle (P1 cell). Timelapse images are shown from S-phase to metaphase. GFP-PSF1 initially persists on prophase chromatin following depletion of LRR-1 (the arrows denote examples), before being released in late prophase (indicated by asterisk). Scale bars correspond to 5µm. (b) The duration of the indicated cell cycle phases for the experiment in (a) were measured as described in Methods. The data are expressed relative to the length of the corresponding period in control embryos, and represent the mean values (n = 5 embryos; the lines on the boundary of each cell cycle phase indicate standard deviations from the mean). (c) Worms homozygous for GFP-psf-1 and lrr-1Δ were grown in parallel to the equivalent heterozygote (control), as described in Methods. After exposure to atl-1 RNAi (this allows homozygous lrr-1Δ germ cells to proceed with meiosis), the resultant embryos were processed as above. The images depict the second embryonic cell cycle (P1 cell), showing persistent association of GFP-PSF-1 with chromatin during prophase (arrows), before release in late prophase (asterisk). (d-e) Homozygous GFP-psf-1 worms were exposed to the indicated RNAi. Embryos were then isolated and processed as in Figure 1e-f. Unprocessed scans of key immunoblots are shown in Supplementary Figure 8.