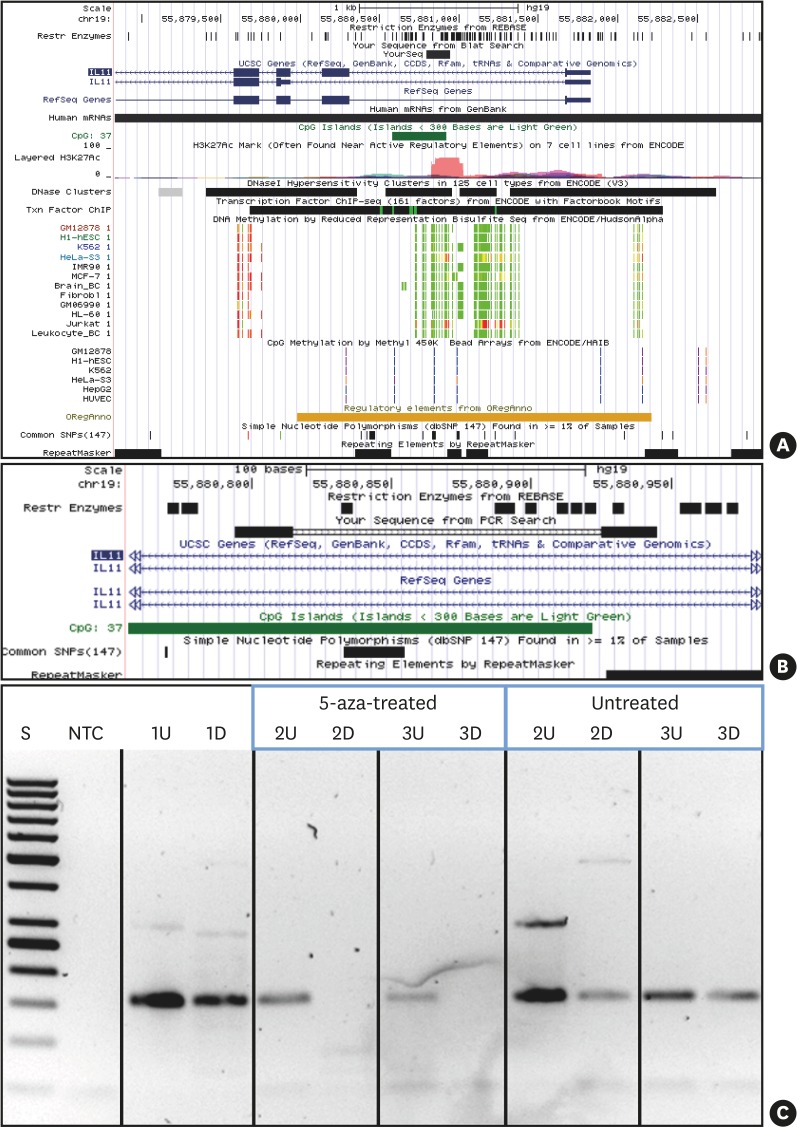
Figure 3.
5-aza treatment causes demethylation of IL11 CpG island methylation. UCSC genome browser (hg19) view indicating the location of the IL11 CpG island and PCR amplicon investigated. (A) IL11 gene region (UCSC genome browser view; hg19) presenting the targeted CpG island (intron 1 within IL11: dark green) and the PCR product (indicated as black bar denoted “YourSeq”). DNA methylation tracks (methyl reduced representation bisulfite sequencing; light green indicated 0% and red indicated 100% methylated CpGs). Only Jurkat and HeLa show methylation of the IL11 CpG island. (B) detailed location of the IL11 CpG island and PCR amplicon (primers, bold black) and location of methylation-sensitive restriction sites (denoted as “Rest Enzymes”). (C) IL11-PCR DNA methylation testing results. 1, leukocyte DNA from a normal healthy individual (not 5-aza-treated); 2 and 3, fibroblast DNA: 5-aza-treated vs. non-treated as indicated.
5-aza: 5-aza-2'-deoxycytidine, CpG: commonly next to a guanosine, UCSC: University of California, Santa Cruz, IL11: interleukin-11, PCR: polymerase chain reaction, U: undigested, D: digested by methylation-sensitive restriction enzymes, S: 50 bp ladder, NTC: non-template PCR control 1.