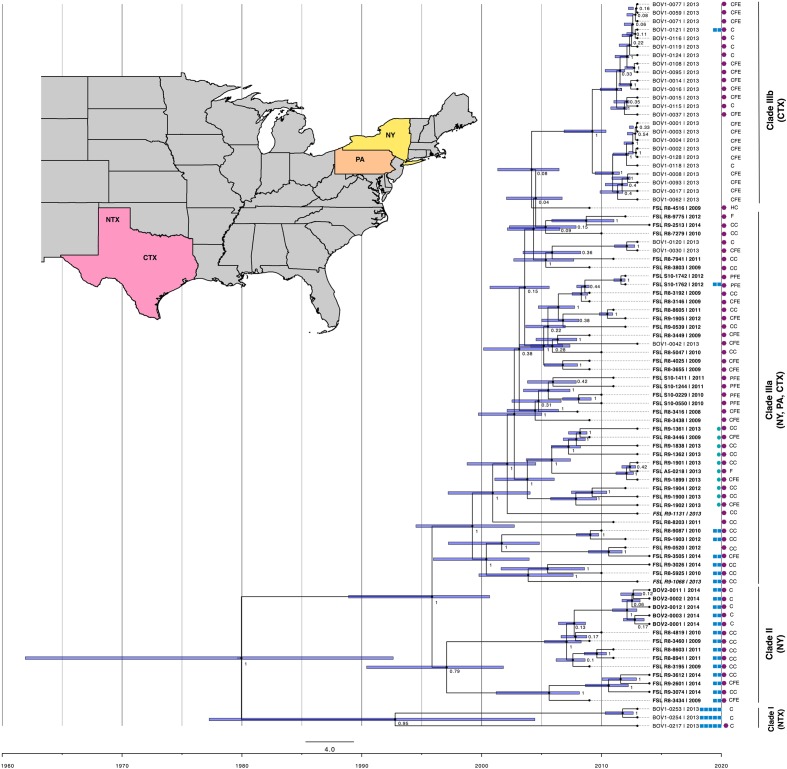
FIGURE 1.
Tip-dated phylogeny of 86 Salmonella enterica subsp. enterica serotype Cerro isolates from NY (isolate names in bold print), PA (isolate names in italic print), CTX (isolate names in regular print) and NTX (isolate names in regular print), isolated from cattle (C; data on clinical symptoms unavailable), cattle with clinical symptoms (CC), cattle farm environment (CFE), produce farm environment (PFE), food (F) and human clinical case (HC). The unrooted tree was constructed based on high quality core genome single nucleotide polymorphisms (SNPs) in BEAST using general time-reversible (GTR) substitution model, strict molecular clock model and coalescent constant size population model, in four independent runs of 100,000,000 Monte Carlo Markov Chain (MCMC) simulations. The violet bars on the nodes represent credible intervals for time of taxa divergence. The cluster of isolates with abolished hydrogen sulfide production is marked with turquoise circles. The blue squares represent from 0 to 5 detected genes in D-alanine transporter-encoding cluster in following order from left to right: STM1633, ST1634, ST1635, STM1636, and STM1637. Purple hexagons represent isolates that carry orgA gene encoding oxygen-regulated invasion protein.