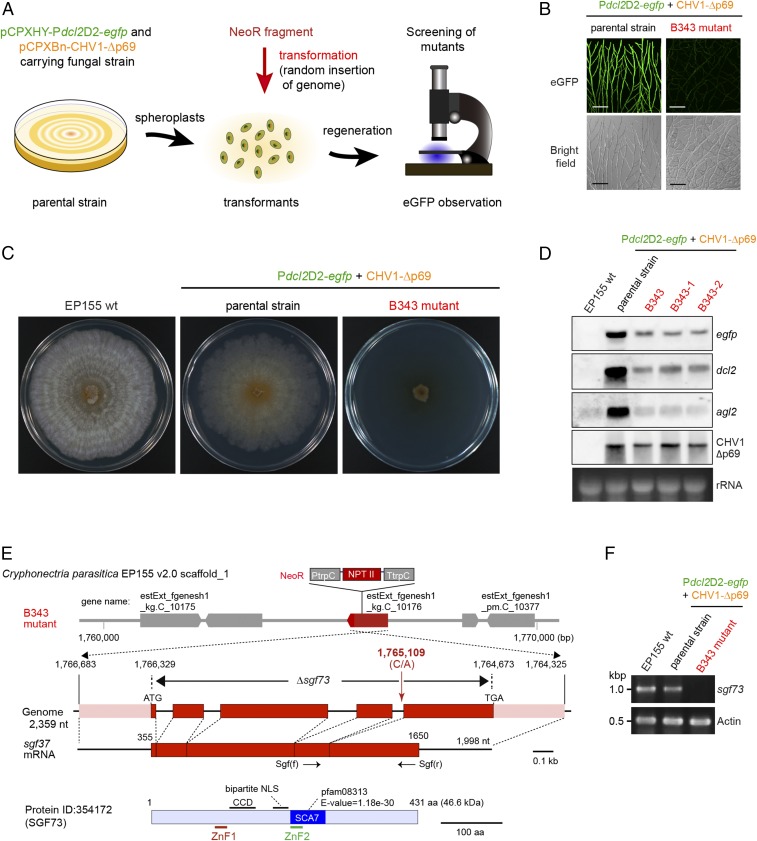
Fig. 2.
Genetic screen for host factors involved in the activation of antiviral RNA silencing in C. parasitica. (A) Screening procedure to identify host genes mediating the activation of antiviral RNA silencing. Protoplasts of the parental fungal strain (EP155/Pdcl2D2-egfp+CHV1-Δp69) (Fig. 1A and Table S1) were randomly mutagenized by introducing a cassette (NeoR) carrying a neomycin phosphotransferase II gene (triple transformation) and screened for loss of eGFP fluorescence under confocal laser scanning microscopy. (B) Loss of GFP induction in a selected mutant, B434. The parental fungal strain green-fluoresced, and the mutant B434 carrying mutagenic construct NeoR failed to do so. (Scale bars, 0.1 mm.) (C) One-week-old PDA cultures of the standard EP155 strain, parental strain (Pdcl2D2-egfp+CHV1-Δp69), and B343. (D) Northern analysis of three fungal strains used in A and independent single spore isolates (B343-1 and B343-2) derived from B343. (E) A diagram of the sgf73 gene and NeoR insertion site in B434. The NeoR is shown by fusion primer and nested integrated-PCR (21) to be inserted into one of the four introns of the sgf73 gene (protein ID, 354172; scaffold no. 1), a putative component of the SAGA complex. (See Fig. S5 for the sgf73 sequences.) Map positions in this and subsequent figures are shown according to the genome sequence of C. parasitica strain EP155. The SGF73 is characterized by the presence of domain SCA7 carrying a Zn finger motif (ZnF2), a second Zn finger motif (ZnF2), and a putative bipartite NLS. An sgf73 disruptant (Δsgf73) shown in the following figure lacks the entire coding region. (F) RT-PCR analysis of sgf73 expression in EP155 and B434. ssRNA fractions from virus-free EP155, parental strain (Pdcl2D2-egfp+CHV1-Δp69) and B434 were used in RT-PCR for detection of sgf73 and actin mRNAs (Table S3 for primer sequences) were used.