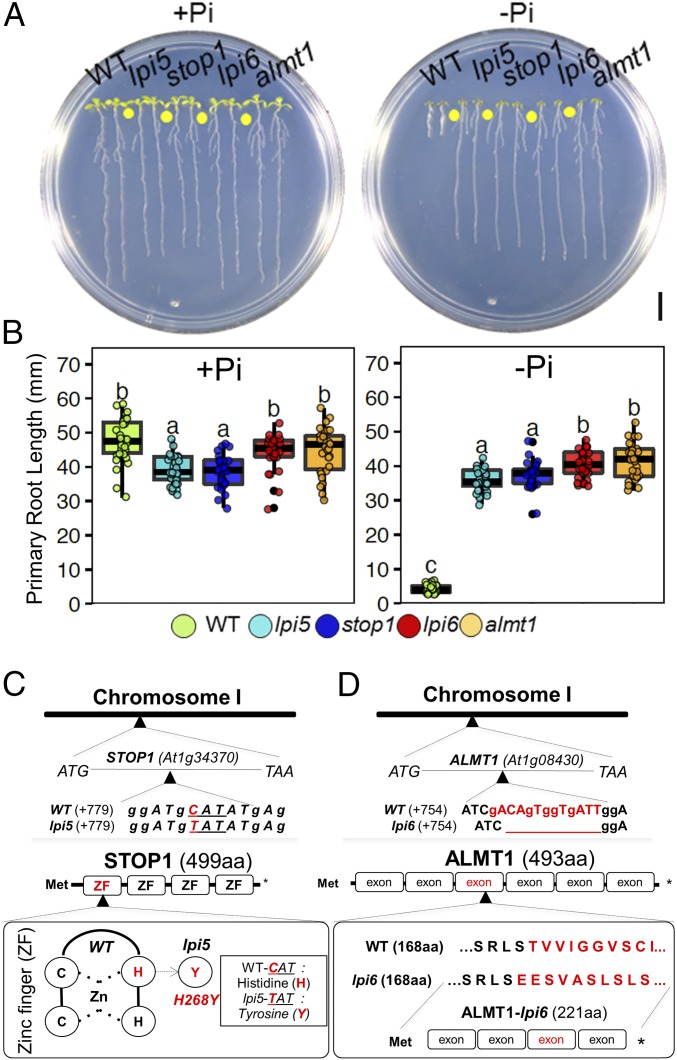
Fig. 2.
Mapping by sequencing revealed lpi5 and lpi6 to be stop1 and almt1 mutants, respectively. (A) Phenotypes of Col-0, lpi5, stop1, almt1, and lpi6 seedlings grown under high-phosphate (+Pi) and low-phosphate (−Pi) conditions 10 dag. (Scale bar, 1 cm.) (B) Primary root length of WT, lpi5, stop1, almt1, and lpi6 seedlings grown under high-phosphate (+Pi) or low-phosphate (−Pi) conditions 10 dag. Dots depict WT, lpi5, stop1, almt1, and lpi6 individuals (n = 30 from three independent experiments). Statistical groups were determined using a Tukey HSD test (P value <0.05) and are indicated with letters. (C) C:T (CAT: TAT) substitution in the +84 base within the At1g34370 locus of the lpi5 genome. lpi5 mutation causes an amino acid substitution (H168Y) that replaces one of the two histidine residues of the four zinc fingers that constitute STOP1. (D) A 13-bp deletion in the +757 base pair position within the first exon of the At1g08430 locus of the lpi6 genome. The 13-bp deletion present in lpi6 causes a frameshift mutation that produces an aberrant protein with 200 amino acids less than the WT. Met, methionine.