Significance
DNA glycosylases are important repair enzymes that safeguard the integrity of the genome by excising chemically damaged DNA bases from the phosphoribose backbone. Recently, these enzymes were found to repair DNA interstrand cross-links (ICLs). ICLs are highly toxic DNA lesions formed by various bifunctional metabolites, environmental toxins, and chemotherapeutic agents that block normal DNA metabolism. This work describes the crystal structure of a newly discovered bacterial DNA glycosylase that repairs ICLs formed by azinomycin B, a potent antimicrobial and antitumor agent. The protein belongs to a structural superfamily prevalent in pathogenic bacteria and may serve as an important therapeutic target.
Keywords: DNA repair, DNA glycosylase, interstrand cross-link, azinomycin B, winged-helix
Abstract
DNA glycosylases are important editing enzymes that protect genomic stability by excising chemically modified nucleobases that alter normal DNA metabolism. These enzymes have been known only to initiate base excision repair of small adducts by extrusion from the DNA helix. However, recent reports have described both vertebrate and microbial DNA glycosylases capable of unhooking highly toxic interstrand cross-links (ICLs) and bulky minor groove adducts normally recognized by Fanconi anemia and nucleotide excision repair machinery, although the mechanisms of these activities are unknown. Here we report the crystal structure of Streptomyces sahachiroi AlkZ (previously Orf1), a bacterial DNA glycosylase that protects its host by excising ICLs derived from azinomycin B (AZB), a potent antimicrobial and antitumor genotoxin. AlkZ adopts a unique fold in which three tandem winged helix-turn-helix motifs scaffold a positively charged concave surface perfectly shaped for duplex DNA. Through mutational analysis, we identified two glutamine residues and a β-hairpin within this putative DNA-binding cleft that are essential for catalytic activity. Additionally, we present a molecular docking model for how this active site can unhook either or both sides of an AZB ICL, providing a basis for understanding the mechanisms of base excision repair of ICLs. Given the prevalence of this protein fold in pathogenic bacteria, this work also lays the foundation for an emerging role of DNA repair in bacteria-host pathogenesis.
The chemical integrity of DNA is constantly challenged by cellular and environmental genotoxic agents that produce a diverse array of covalent nucleobase adducts. If left unrepaired, these DNA lesions impair important cellular processes, including replication, transcription, and cell cycle regulation, leading to mutations, chromosomal rearrangements, and genomic instability that threaten the livelihood of the organism and lead to human diseases (1). Small adducts are typically removed by base excision repair (BER), in which a lesion-specific DNA glycosylase catalyzes hydrolysis of the N-glycosidic bond of the aberrant nucleobase, liberating it from the phosphoribose backbone (2). The resulting abasic site is nicked at the 5′ side by an apurinic/apyrimidinic (AP) endonuclease to create a free 3′-hydroxyl group necessary for the synthesis of new DNA. In contrast, bulky and helix distorting lesions are removed by nucleotide excision repair (NER), which involves removal of an oligonucleotide segment containing the lesion through the action of dual nuclease incisions, followed by helicase removal of the damaged segment.
Interstrand cross-links (ICLs) from various endogenous and environmental sources covalently tether the opposite strands of DNA and pose a major obstacle to normal DNA metabolism (3, 4). The cytotoxicity of cross-linking agents makes them particularly effective as antitumor drugs (5). Because ICLs damage both strands of DNA, repair of ICLs is more elaborate than repair of monoadducts and involves multiple pathways (4, 6). In ICL repair, nuclease-dependent dual incisions on one strand flanking the lesion produce a gapped intermediate that must be filled by translesion synthesis (TLS) or homologous recombination (HR) before NER-associated repair of the remaining unhooked monoadduct can occur.
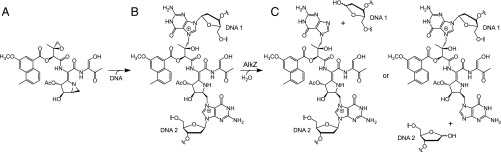
Azinomycin B (AZB) (Fig. 1A) is a cytotoxic, nonribosomal peptide-polyketide secondary metabolite of Streptomyces sahachiroi and Streptomyces griseofuscus (7, 8). This bifunctional alkylating agent forms ICLs in vitro and in vivo (9) and displays potent antibiotic and antitumor activities comparable to the chemotherapeutic mitomycin C (8, 10, 11). AZB ICLs are formed between the electrophilic aziridine and epoxide functional groups of AZB and the N7 nitrogens of guanosine and adenosine within d(GNC) and d(GNT) sequences (Fig. 1B) (12, 13). Computational modeling of the AZB ICL structure is consistent with the entire AZB molecule residing in the major groove of the DNA without nucleobase intercalation by the naphthalate moiety (14, 15).
Fig. 1.
Unhooking of AZB ICLs by AlkZ. (A) AZB. (B) AZB cross-links opposing guanines at their N7 nitrogens via reactive aziridine and epoxide groups. (C) AlkZ glycosylase catalyzes hydrolysis of the N-glycosidic bond of either cross-linked dG to produce two possible dG-AZB-Gua monoadducts.
In contrast to NER-associated repair of other ICLs, AZB ICLs were recently identified as the target of repair by a DNA glycosylase (16). The azi36 (orf1) gene sits adjacent to the AZB synthesis cluster in S. sahachiroi (17) and provides self-resistance to cells from AZB toxicity. The orf1 gene product unhooks AZB ICLs by cleaving the N-glycosidic bond on at least one side of the lesion to produce an abasic site that is recognized by the bacterial AP endonuclease EndoIV (Fig. 1C) (16). Orf1 belongs to the HTH_42 (Pfam 06224) superfamily of uncharacterized winged helix (WH)-containing proteins that exist in various pathogenic and antimicrobial-producing bacteria. However, there are no known structures of any HTH_42 protein, and thus the molecular rationale for glycosylase repair of AZB ICLs is unclear.
The significance of this alternative, glycosylase-mediated ICL repair pathway is underscored by the recent discovery that the unrelated eukaryotic NEIL3 glycosylase unhooks psoralen and abasic site-adenine ICLs during DNA replication (18), and that human NEIL1 can excise unhooked psoralen monoadducts and ICLs (19, 20). Thus, glycosylase-mediated ICL repair activity has been identified in both prokaryotes and eukaryotes by two distinct proteins. The molecular details of how these enzymes can excise adducts covalently tethered to both DNA strands is unknown, however. To better understand the mechanisms for DNA glycosylase-mediated ICL repair, we determined the crystal structure of the S. sahachiroi azi36 gene product and identified the active site by mutational analysis. The structure is the defining member of the HTH_42 superfamily and an eighth structural class of DNA glycosylase. Our analysis shows how the HTH_42 architecture can support unhooking of either side of an AZB ICL or excision of an AZB-dG monoadduct. To better reflect the activity of the enzyme for base excision of alkylated DNA, we propose renaming the Azi36/Orf1 protein “AlkZ.”
Results
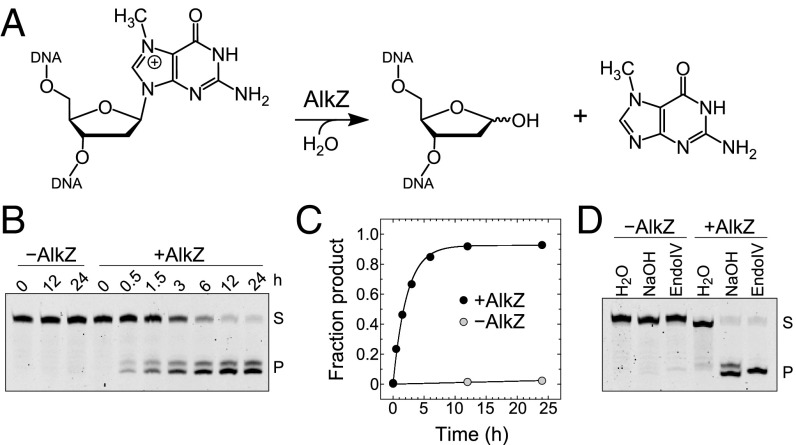
AlkZ Excises N7-Methylguanine from DNA.
Recently, the AlkZ protein was shown to act on AZB ICLs and monoadducts through hydrolysis of one or more of the N-glycosidic bonds (16), but whether the enzyme would act on adducts other than AZB was unclear. DNA glycosylases that excise alkylated DNA lesions often exhibit activity for a number of N3- and N7-methylated purines in addition to their preferred substrates (21, 22). For example, Bacillus cereus AlkD displays robust activity for bulky minor groove N3-yatakemycinyldeoxyadenosine lesions, but also cleaves N3-methyldeoxyadenosine and N7-methyldeoxyguanosine (d7mG) (23, 24). Therefore, we tested the ability of AlkZ to catalyze excision of N7-methylguanine (7mGua) from an oligonucleotide containing d7mG (Fig. 2A).
Fig. 2.
Excision of N7-methylguanine by AlkZ. (A) Base excision of d7mG by AlkZ. (B) Denaturing PAGE of d7mG-DNA substrate (S) and nicked abasic-DNA product (P) after treatment without (−) or with (+) AlkZ for the specified time, followed by alkaline hydrolysis. (C) Quantitation of the gel shown in B, averaged across three independent experiments. Error bars represent SD. (D) Denaturing PAGE of d7mG-DNA substrate (S) and nicked abasic-DNA product (P) after treatment without (−) or with (+) AlkZ for 24 h, followed by treatment with water, NaOH, or EndoIV.
Using a standard in vitro assay that measures glycosylase activity by alkaline hydrolysis of the resulting abasic site product, we found that AlkZ hydrolyzed d7mG with an observed rate (kobs) of (8.5 ± 0.4) × 10−5 s−1 (Fig. 2 B and C). This d7mG activity is modest compared with that of other alkylpurine DNA glycosylases under similar conditions (21, 25, 26), but demonstrates that an AZB adduct is not necessary for AlkZ activity and corroborates the previous data showing activity toward monoadducts (16). In addition, we found that treatment with NaOH or EndoIV is required to nick the abasic DNA produced by AlkZ, indicating that AlkZ is a monofunctional DNA glycosylase that does not have lyase activity (Fig. 2D).
AlkZ Adopts a Unique DNA-Binding Fold.
HTH_42 superfamily proteins are predicted to contain two WH motifs, which are common helix-turn-helix (HTH)-related DNA-binding elements found in transcription factors and DNA processing enzymes (27). The canonical WH fold contains a three-helix bundle at the N terminus and a three-stranded β-sheet (wing) at the C terminus, and is related to the HTH sequence-specific DNA-binding motif in that α3 serves as a “recognition helix” by inserting into the DNA major groove (28). However, because there is no known structure of an HTH_42 protein, how or if the putative WH motifs bind DNA remains unclear. Moreover, there are no WH motifs in any of the seven known DNA glycosylase protein folds (29), so the molecular details of how AlkZ acts as a DNA repair enzyme are also unclear.
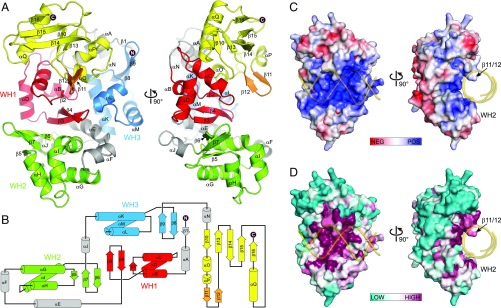
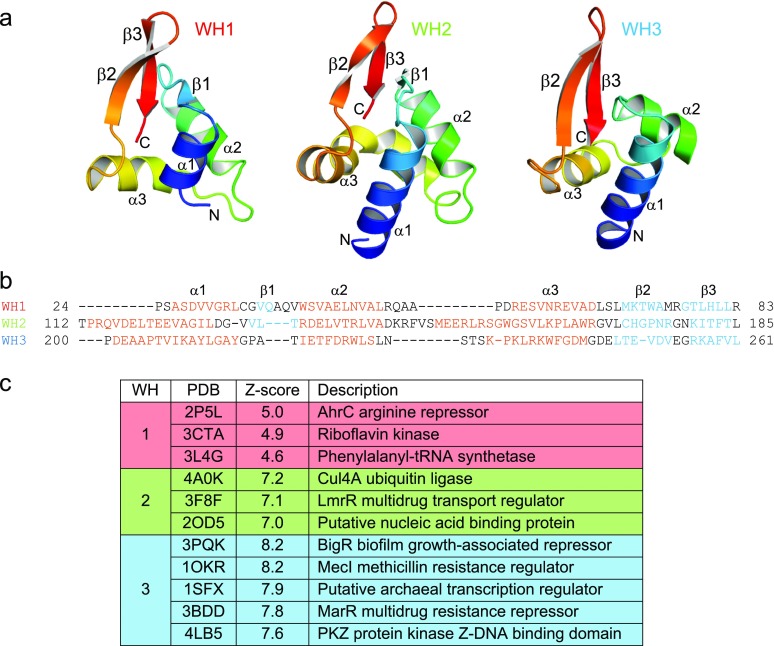
To understand the structural basis for DNA glycosylase-mediated ICL repair by AlkZ, we determined the crystal structure of the S. sahachiroi enzyme. Experimental X-ray phases were determined from single-wavelength anomalous dispersion using selenomethionyl-substituted protein (Fig. S1). The resulting crystallographic model, which contains all 371 residues of the protein, was refined against X-ray diffraction data extending to 2.3 Å, resulting in an R factor of 15.7% and an Rfree value of 20.1% (Table S1). The protein adopts a cluster of three tandem WH motifs (WH1, WH2, and WH3) that pack against a C-terminal β-barrel subdomain to form a C-shaped molecule (Fig. 3). WH1 is centrally located and bridges the WH2 and WH3 motifs. WH1 packs against WH2 with twofold rotational symmetry and their β-wings at the interface, and against WH3 in a 90°-offset head-to-tail arrangement. This cluster of WH motifs appears to be a unique protein architecture, as a search for structural homologs using the DALI server (30) returned no results aside from those of each individual WH (Fig. S2). Consistent with the established structural diversity among WH motifs, the three motifs in AlkZ show slight variations from one another (Fig. S2), including a highly kinked α3 “recognition helix” in WH2 (helix αI). However, with the exception of WH3, which lacks the short β1 strand, each WH adopts the canonical α1-β1-α2-α3-β2-β3 topology.
Fig. S1.
Crystallographic data. Stereoimages of helix αG (residues T112–L127) superimposed against experimental Se-SAD (A) and refined 2Fo−Fc (B) electron density contoured at 1σ.
Table S1.
X-ray data collection and refinement statistics
| Statistic | Value | |
| Data collection | ||
| Space group | P65 | |
| Cell dimensions | ||
| a, b, c, Å | 78.92, 78.92, 139.40 | |
| α, β, γ, ° | 90.00, 90.00, 120.00 | |
| Resolution, Å | 50.00–2.30 (2.38–2.30)* | |
| Rsym | 0.118 (0.481) | |
| Avg. I/σI | 29.1 (7.0) | |
| Completeness, % | 96.2 (100.0) | |
| Redundancy | 12.3 (12.6) | |
| Wilson B-factor, Å2 | 21.8 | |
| Refinement | ||
| Resolution, Å | 39.47–2.30 (2.42–2.30) | |
| No. of reflections | 20,828 (3,014) | |
| Rwork | 0.157 (0.170) | |
| Rfree† | 0.201 (0.220) | |
| No. of atoms‡ | ||
| Protein | 2,882 | |
| Water | 245 | |
| Avg. B-factors‡,§, Å2 | ||
| Protein | 25.0 | |
| Water | 27.6 | |
| rmsd | ||
| Bond lengths, Å | 0.003 | |
| Bond angles, ° | 0.769 | |
| Ramachandran distribution, % | ||
| Favored | 98.1 | |
| Allowed | 1.9 | |
| Disallowed | 0.0 | |
Statistics for the highest-resolution shell are shown in parentheses.
Rfree was determined from the 5% of reflections excluded from refinement.
Riding hydrogens were not included in the number of atoms or average B-factors.
Equivalent isotropic B-factors were calculated in conjunction with TLS-derived anisotropic B-factors.
Fig. 3.
Crystal structure of AlkZ. (A) Orthogonal views of the enzyme colored by subdomain. WH motifs are in red, green, and blue; the C-terminal domain is in yellow; the putative DNA-binding β11/12 loop is in orange; and connecting helices are in gray. (B) Topology diagram color-coded as in A. (C) Solvent-accessible surface of AlkZ colored by electrostatic potential (−7 to +7 kBT/eC). (D) Solvent-accessible surface of AlkZ colored by sequence conservation. In C and D, B-DNA modeled against the crystal structure is shown as a transparent gold backbone trace.
Fig. S2.
Structural homology of AlkZ WH motifs. (A) Comparison of WH structures, colored rainbow from amino (blue) to carboxy termini (red). Secondary structural elements are named using WH convention. (B) Structure-based sequence alignment of WH domains, with α-helical regions in red and β-sheet regions in blue. (C) Dali results for structures of individual AlkZ WH motifs.
The most notable feature of the C-shaped structure is the concave channel that spans the width of the protein. The channel is defined by all three WH motifs and a short β-hairpin (β11/12) that protrudes from the C-terminal domain. The surface of this channel is lined with positively charged, highly conserved residues and is appropriately sized to accommodate duplex DNA (Fig. 3 C and D). DNA bound along this channel would be clamped between the β11/12 hairpin on one side of the duplex and helix αI—the putative “recognition helix” of WH2—on the other side, indicating that WH2 may play a role in DNA recognition similar to that of other WH motifs. In contrast, the corresponding helices from WH1 (αD) and WH3 (αM) reside outside the central channel in positions not likely to bind DNA. Indeed, docking DNA from other WH-DNA and HTH-DNA complexes onto WH1 or WH3 positioned the DNA well outside of the putative DNA-binding channel.
Active Site Residues Reside Within the Putative DNA-Binding Channel.
To gain insight into how AlkZ properly orients an AZB lesion for catalysis, we docked a B-DNA model containing a computationally derived AZB ICL (14) as a rigid body into the putative DNA-binding channel (Fig. 4 A–D). Two nearly equivalent DNA orientations, related by the symmetric nature of the AZB ICL relative to the DNA dyad axis, optimized van der Waals and electrostatic interactions without altering the structure of the protein or the DNA. Both orientations placed the β11/12 hairpin into the minor groove directly across from the AZB ICL and placed WH2 against the adduct in the major groove.
Fig. 4.
Identification of the AlkZ active site. (A and B) Orthogonal views of the AlkZ active site with AZB-ICL-DNA docked in the putative DNA-binding channel. The protein is color-coded as in Fig. 3, and AlkZ residues tested for their roles in base excision activity are labeled. The AZB-ICL-DNA model is shown as a slate/charcoal backbone trace with adducted guanosines (slate/charcoal) and AZB (purple) depicted in ball-and-stick representation. (C and D) Orthogonal views of AlkZ bound to AZB-ICL-DNA in a hypothetical alternate binding orientation. (E) Base excision activity of wild-type (WT) and mutant AlkZ proteins as measured by the formation of nicked abasic-DNA product (P) after incubation of protein or buffer (mock) with d7mG-DNA substrate (S) for 24 h.
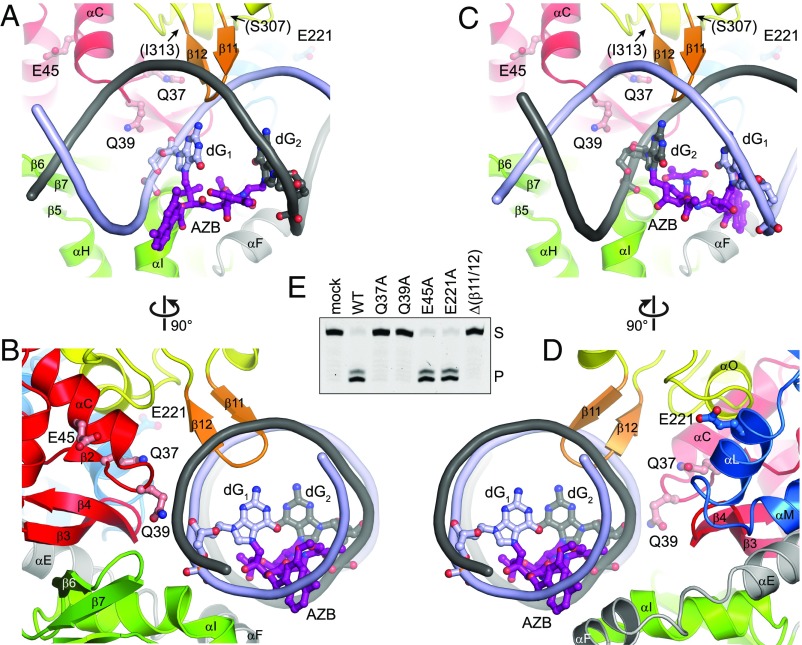
We used these models to identify candidate catalytic residues among the highly conserved polar, cationic, aromatic, and hydrophobic side chains lining the concave surface (Fig. S3A), and thus likely in the vicinity of a bound DNA lesion. Monofunctional glycosylases generally initiate nucleophilic attack from the minor groove using carboxylate (Asp, Glu) or carboxamide (Asn, Gln) side chains (31, 32). In AlkZ, side chains from conserved glutamines Q37 and Q39 (Fig. S4) point into the minor groove at either point of ICL attachment in both orientations of our docking model (Fig. 4 A–D). Whereas Q37 is recessed at the rear of the channel, Q39 protrudes far enough into the minor groove to orient a water nucleophile for catalysis.
Fig. S3.
AlkZ active site and mutants. (A) The region containing catalytic residues in AlkZ shown as a stereoimage and color-coded as in Fig. 3. (B) SDS/PAGE of purified proteins used in biochemical studies. (C) Thermal denaturation as monitored by circular dichroism. Spectra were normalized from minimum and maximum dichroism values. Melting temperatures (Tm) were derived by fitting the data to the equation θ = 1/(1 + e(Tm − Tk)).
Fig. S4.
AlkZ sequence conservation. The sequence of S. sahachiroi AlkZ is aligned against seven orthologs chosen at random from the top 250 hits from a PSI-BLAST search. The secondary structure deduced from the crystal structure is shown at the top. Triangles designate Q37 and Q39 residues essential for catalysis (black), residues lining the concave active site cleft (gray), and residues predicted to contact DNA (white).
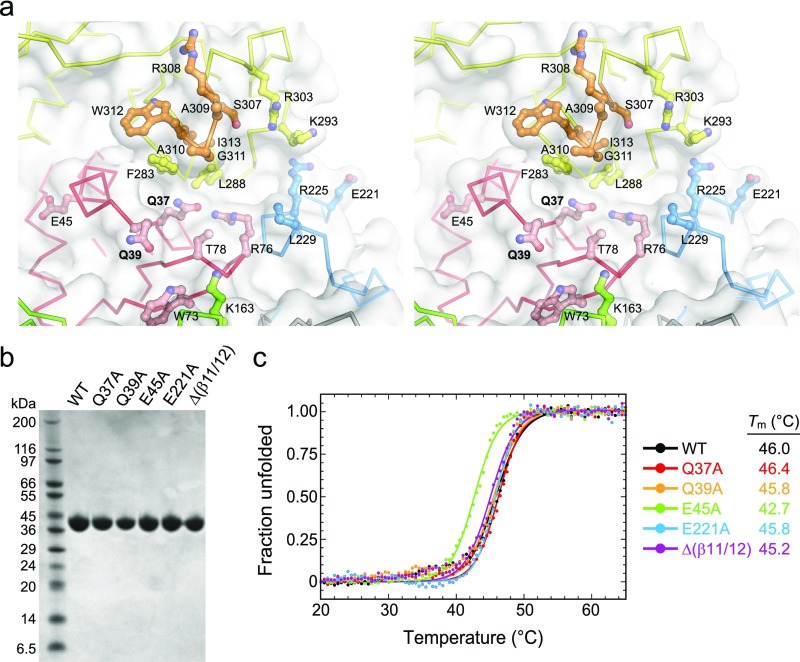
We tested involvement of the helix-penetrating QxQ motif and β11/12 hairpin in enzyme activity by substituting with alanine or glycine and measuring d7mG excision activity from purified proteins (Fig. 4E). We verified by circular dichroism that all mutant proteins were folded under the conditions of our assay (Fig. S3 B and C). Both Q37A and Q39A mutants completely abrogated d7mG excision, even after 24 h. In contrast, alanine substitution of the nearest glutamate side chains, E45 and E221, which reside just outside the putative active site on helices αC (WH1) and αL (WH3), respectively, had no effect on base excision activity. In addition to catalysis by polar residues, DNA glycosylases often stabilize a particular DNA conformation in the enzyme-substrate complex by inserting a steric wedge—either bulky side chains or a loop—into the DNA helix. In AlkZ, this role is likely played by the β11/12 hairpin, which protrudes into the minor groove in our docking model (Fig. 4 A–D). Consistent with this hypothesis, replacing residues 307–313 that form the β11/12 loop with two glycine residues (Δβ11/12) abrogated d7mG excision activity (Fig. 4E). These results identify this central region of the protein as the active site, with important roles for Q37, Q39, and the β11/12 hairpin in base excision.
Discussion
This work defines the architecture of the HTH_42 family of DNA-binding proteins (33) and an eighth class of DNA glycosylase (29, 34). A unique cluster of three tandem WH motifs scaffold a putative DNA-binding channel, with only one WH (WH2) positioned to participate in the canonical helix-major groove recognition used by other WH and HTH motifs (28). The active site contains a QxQ motif and a β-hairpin essential for base excision activity and conserved among AlkZ homologs and the HTH_42 superfamily (Fig. S4). Our docking models predict insertion of the β11/12 hairpin into the DNA minor groove at the lesion site. This motif is likely involved in stabilizing a particular DNA conformation in the enzyme-substrate complex, similar to that observed for β-hairpins and loops in other glycosylases (29). The N-terminal glutamine (Q37) in the QxQ motif is receded in the active site cleft and thus predicted to bind the DNA backbone, consistent with histidine occupying this position in some AlkZ homologs. The C-terminal glutamine (Q39), on the other hand, likely plays a catalytic role. Our docking models predict that Q39 protrudes into the minor groove so that its carboxamide side chain is positioned to orient a water nucleophile in the manner observed for N140 in human thymine DNA glycosylase (32) and for aspartate or glutamate side chains in other glycosylases (31). Consistent with such a catalytic role for Q39, approximately one-half of all HTH_42 proteins have an aspartate at this position, suggesting that at least some other members of this previously uncharacterized superfamily also have DNA glycosylase activity.
AlkZ is one of two DNA glycosylases that unhooks an ICL (16, 18). The mechanism by which a DNA glycosylase unhooks an ICL is not immediately obvious, because these enzymes typically capture the modified nucleoside inside the active site by extruding it from the DNA helix, and such a base-flipping mechanism would be inhibited by an ICL. However, we recently discovered a non–base-flipping mechanism that enables glycosylase excision of bulky minor groove adducts and conceivably could enable ICL excision. The bacterial AlkD glycosylase is able to recognize and cleave deoxyadenosine adducts of the bulky natural product yatakemycin (YTM) without rotating the lesion from the duplex (24). Like AlkZ, AlkD adopts a C-shaped fold that engages DNA along the concave surface (22). The AlkD active site positions a catalytic aspartate and two catalytic tryptophan side chains (25) against the deoxyribose of the alkylated adenosine, which is displaced only slightly into the minor groove of the DNA. Similarly, in our AlkZ docking models, the QxQ motif is positioned sufficiently far into the minor groove to access the N-glycosidic bonds of the ICL without rotating the cross-linked nucleosides (Figs. 4 and 5). The eukaryotic DNA glycosylase with ICL unhooking activity, NEIL3, also has an active site (35) that conceivably could access an unflipped lesion.
Fig. 5.
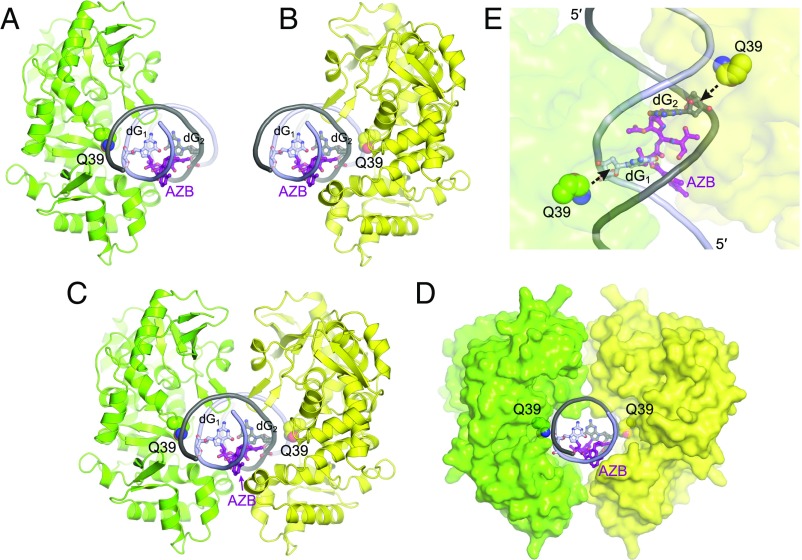
Possible mechanisms of ICL repair by AlkZ. (A and B) Two binding orientations of AlkZ against each side of an AZB ICL. The catalytic Q39 side chain is shown in green or yellow, and the AZB ICL is shown in purple. (C and D) Dimeric model of two AlkZ subunits bound to one AZB ICL. The proteins are shown as green and yellow cartoons (C) or solvent-accessible surfaces (D). (E) Top view of the hypothetical AlkZ dimer interface, showing simultaneous engagement of both cross-linked dG residues by the catalytic Q39 side chain.
Previous work has established that AlkZ can unhook an AZB ICL, but whether the enzyme cleaves both sides is less clear (16). Although full ICL excision by a DNA glycosylase would create opposing abasic sites susceptible to a deleterious double-strand break, it also would simplify repair by eliminating the need for NER or TLS/HR pathways to remove the remaining monoadduct. Pol I and II bypass of abasic sites and major groove monoadducts (36, 37) suggests that, in principle, BER could fully process opposing abasic sites in bacteria. Several lines of evidence suggest that AlkZ can excise the entire cross-link by one of two mechanisms, either by sequentially unhooking the ICL and excising the remaining monoadduct or by simultaneously cleaving the glycosidic bonds of both modified dG residues. First, AlkZ has both unhooking activity for ICLs and excision activity for monoadducts (Fig. 2 and ref. 16). Second, our DNA docking models illustrate that AlkZ can bind to either side of the AZB ICL (Fig. 5 A and B). Both binding orientations position the putative active site against the minor groove on either side of the ICL without obstruction from AZB. Third, our docking models show that the two binding orientations can coexist at the same ICL. Modeling two AlkZ molecules on opposite sides of the DNA with their respective active sites against the two cross-linked dG residues reveals a remarkable complementarity between protein surfaces (Fig. 5 C–E). Although we have no evidence of dimerization of AlkZ in the presence of d7mG-DNA, dimerization may be dependent on the AZB ICL. The recent discovery of a dimeric DNA glycosylase acting as part of a restriction-modification system in thermophilic bacteria (34) raises the possibility that AlkZ could use a similar strategy to simultaneously excise the entire cross-link. However, at the lower temperatures at which AlkZ functions, spontaneous formation of a double-strand break from opposing abasic sites would be less likely. Moreover, the bound AlkZ dimer may transiently shelter both abasic sites from hydrolysis until further repair by enzymes in the BER pathway occurs.
Pathogenic and antibiotic-producing microbes have resistance mechanisms against their own toxins that often are genetically clustered with the antibiotic synthesis operon (38–40). AlkZ is the second DNA glycosylase discovered to act as a toxin resistance mechanism (16). The AlkD homolog YtkR2 is produced in Streptomyces sp. TP-A0356 as a means to protect against its natural product YTM, a bulky genotoxin of the spirocyclopropylcyclohexadienone family (41, 42). Similar to the AlkZ gene (azi36/orf1), ytkR2 and putative BER genes are embedded within the YTM synthesis gene cluster. Although AlkZ and YtkR2 likely originated in AZB- and YTM-producing organisms, homologous proteins are evident in diverse bacteria, including human pathogens (16, 43). Thus, the two bacterial DNA glycosylases known to repair bulky and cross-linked DNA damage arising from potent bacterial toxins have transferred among bacterial populations. This implicates DNA repair and alternative mechanisms thereof, in possible treatments against antibiotic resistance, and opens the possibility that other DNA repair mechanisms and genotoxic metabolites await discovery in pathogenic and antibiotic-producing bacteria.
Methods
Protein Purification.
The azi36 gene was amplified by PCR from genomic DNA isolated from S. sahachiroi (American Type Culture Collection; 33158) and cloned into pBG102 (Vanderbilt University Center for Structural Biology). His6-SUMO-AlkZ was overexpressed in Escherichia coli Tuner(DE3) cells at 16 °C for 18 h in LB medium supplemented with 100 µg/mL kanamycin and 50 µM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cells were lysed with gentle sonication, and cell debris was removed by centrifugation at 45,000 × g at 4 °C for 30 min. Clarified lysate was passed over 5 mL of Ni-NTA agarose resin equilibrated in buffer A [50 mM Tris⋅HCl pH 8.5, 500 mM NaCl, and 10% (vol/vol) glycerol]. The column was washed with 100 mL of 20 mM imidazole/buffer A, and protein was eluted in 50 mL of 250 mM imidazole/buffer A. Protein fractions were pooled and supplemented with 0.1 mM EDTA and 1 mM tris(2-carboxyethyl)phosphine (TCEP) before incubation with ∼0.5 mg of Rhinovirus 3C protease (PreScission) at 4 °C overnight. Cleaved protein was diluted 10-fold in buffer B [50 mM Tris⋅HCl pH 8.5, 10% (vol/vol) glycerol, 1 mM TCEP, and 0.1 mM EDTA]. Diluted protein was loaded onto a 5-mL heparin Sepharose column, washed with 25 mL of 50 mM NaCl/buffer B, and eluted with 50 mL of a 50–1,000 mM NaCl/buffer B linear gradient. Fractions were pooled and repassed over Ni-NTA agarose resin equilibrated in buffer A. Flow-through and protein that eluted with 50 mL of 20 mM imidazole/buffer A were pooled, concentrated and filtered, and passed over Superdex 200 size exclusion resin equilibrated in buffer C [20 mM Tris⋅HCl pH 8.5, 100 mM NaCl, 5% (vol/vol) glycerol, 1 mM TCEP, and 0.1 mM EDTA]. Protein was eluted with 1.5 CV of buffer C, concentrated to 6 mg/mL, flash-frozen in liquid nitrogen, and stored at −80 °C.
AlkZ mutants were generated using the Q5 Mutagenesis Kit (New England BioLabs). The Δ(β11/12) mutant was generated by replacing residues 307-SRAAGWI-313 with two glycine residues. Mutant proteins were overexpressed and purified the same as wild-type (WT). The structural integrity of mutant proteins was verified by thermal denaturation using far-UV circular dichroism spectroscopy, in which molar ellipticity at 222 nm was monitored using a Jasco J-810 spectropolarimeter equipped with a Peltier temperature controller as a solution containing 5 µM enzyme, 50 mM Hepes pH 8.5, 100 mM KCl, and 10% (vol/vol) glycerol was heated at 1 °C/min in a 0.1-cm cell (Fig. S3 B and C). Selenomethionyl-substituted AlkZ (SeMet-AlkZ) was overexpressed and purified the same as the WT protein, except that protein was overexpressed in M9 medium supplemented with 0.4% (wt/vol) dextrose, 1 mM MgSO4, 0.1 mM CaCl2, 1 mg/L thiamine, 70 mg/L selenomethionine, 50 mg/L leucine, 50 mg/L isoleucine, 50 mg/L valine, 100 mg/L phenylalanine, 100 mg/L lysine, 100 mg/L threonine, 100 µg/mL kanamycin, and 50 µM IPTG.
Crystallization, X-Ray Data Collection, and Structure Refinement.
SeMet-AlkZ was crystallized by sitting drop vapor diffusion at 21 °C by mixing equal volumes of 2 mg/mL protein and reservoir solution containing 18% (wt/vol) PEG 8000, 10 mM Tris⋅HCl pH 7.5, and 10 mM MgCl2. Crystals were harvested 4 d after setting the drops, cryoprotected in reservoir solution supplemented with 15% (vol/vol) glycerol, and flash-frozen in liquid nitrogen. X-ray diffraction data from a single frozen crystal were collected at a wavelength of 0.97857 Å at the Advanced Photon Source Beamline 21-ID-G at Argonne National Laboratory and processed with HKL2000 (44). Phasing and refinement were carried out using the PHENIX suite of programs (45). Phases were calculated from the positions of eight Se atoms and modified by solvent flattening using PHENIX AutoSol. A partial model consisting of 347 of 371 SeMet-AlkZ residues was built automatically by PHENIX AutoBuild; the remaining residues were built manually in Coot (46). After adding riding hydrogens with PHENIX ReadySet, atomic coordinates and TLS-derived anisotropic B-factors were refined using PHENIX Refine. The final model was validated with MolProbity (47) and contained no residues in the disallowed regions of the Ramachandran plot. Refinement and validation statistics are presented in Table S1. Atomic coordinates and structure factors were deposited in the Protein Data Bank (PDB) under accession code 5UUJ. The AlkZ-DNA model was prepared by extending the previously determined AZB-ICL-DNA model (14) with three B-form base pairs on each end, followed by manual docking onto the AlkZ structure using PyMOL. All structural biology software was curated by SBGrid (48).
Base Excision Assay.
DNA substrates containing a single d7mG lesion and a 5′-6-carboxyfluorescein (FAM) label within the sequence FAM-d(CACCACTACACC(7mG)ATTCCTTACAAC)/d(GTTGTAAGGAATCGGTGTAGTGGTG) were prepared as described previously (23). In 60-μL reactions, 10 µM AlkZ was incubated with 100 nM FAM-DNA in glycosylase buffer [50 mM Hepes pH 8.5, 100 mM KCl, 10 mM EDTA, and 10% (vol/vol) glycerol] at 25 °C. At various times, 8-µL aliquots were added to 2 µL of 1 M NaOH and heated at 70 °C for 2 min. To test for AP lyase activity, the 8-µL aliquots were added to 2 µL of glycosylase buffer, 1 M NaOH, or E. coli EndoIV (New England BioLabs). Samples were denatured at 70 °C for 5 min in 5 mM EDTA pH 8.0, 40% (wt/vol) formamide, 0.5 mg/mL bromophenol blue, and 0.5 mg/mL xylene cyanol, and electrophoresed on a 20% (wt/vol) acrylamide/8 M urea sequencing gel at 20 W for 2 h in 0.5× TBE buffer (45 mM Tris, 45 mM borate, and 1 mM EDTA pH 8.0). Gels were imaged on a Typhoon Trio variable mode imager (GE Healthcare) using 532-nm excitation and 526-nm emission fluorescence, and bands were quantified with ImageQuant (GE Healthcare). Reactions were performed in triplicate.
Acknowledgments
We thank Stefano Alcaro for providing the AZB-ICL-DNA model and Joshua Bland for assisting with the production of mutant proteins. This work was funded by the National Science Foundation (Grant MCB-1517695) and the National Institutes of Health (Grant R01 ES019625). Support for the Vanderbilt Robotic Crystallization Facility was provided by the National Institutes of Health (Grant S10 RR026915). Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy Office of Science by the Argonne National Laboratory, was supported by the US Department of Energy (Grant DE-AC02-06CH11357). The use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). E.A.M. and G.M.W. were supported by the Vanderbilt Training Program in Environmental Toxicology (Grant T32 ES07028).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5UUJ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703066114/-/DCSupplemental.
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krokan HE, Bjørås M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schärer OD. DNA interstrand crosslinks: Natural and drug-induced DNA adducts that induce unique cellular responses. ChemBioChem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- 4.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajski SR, Williams RM. DNA cross-linking agents as antitumor drugs. Chem Rev. 1998;98:2723–2796. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- 6.Clauson C, Schärer OD, Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Biol. 2013;5:a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hata T, et al. Carzinophilin, a new tumor inhibitory substance produced by streptomyces. I. J Antibiot (Tokyo) 1954;7:107–112. [PubMed] [Google Scholar]
- 8.Nagaoka K, et al. Azinomycins A and B, new antitumor antibiotics. I. Producing organism, fermentation, isolation, and characterization. J Antibiot (Tokyo) 1986;39:1527–1532. doi: 10.7164/antibiotics.39.1527. [DOI] [PubMed] [Google Scholar]
- 9.Terawaki A, Greenberg J. Effect of carzinophillin on bacterial deoxyribonucleic acid: formation of inter-strand cross-links in deoxyribonucleic acid and their disappearance during post-treatment incubation. Nature. 1966;209:481–484. doi: 10.1038/209481a0. [DOI] [PubMed] [Google Scholar]
- 10.Shimada N, et al. Clinical studies of carzinophilin, an antitumor substance. J Antibiot (Tokyo) 1955;8:67–76. [PubMed] [Google Scholar]
- 11.Ishizeki S, et al. Azinomycins A and B, new antitumor antibiotics. III. Antitumor activity. J Antibiot (Tokyo) 1987;40:60–65. doi: 10.7164/antibiotics.40.60. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong RW, Salvati ME, Nguyen M. Novel interstrand cross-links induced by the antitumor antibiotic carzinophilin/azinomycin B. J Am Chem Soc. 1992;114:3144–3145. [Google Scholar]
- 13.Fujiwara T, Saito I, Sugiyama H. Highly efficient DNA interstrand crosslinking induced by an antitumor antibiotic, carzinophilin. Tetrahedron Lett. 1999;40:315–318. [Google Scholar]
- 14.Alcaro S, Coleman RS. A molecular model for DNA cross-linking by the antitumor agent azinomycin B. J Med Chem. 2000;43:2783–2788. doi: 10.1021/jm990362l. [DOI] [PubMed] [Google Scholar]
- 15.Coleman RS, Perez RJ, Burk CH, Navarro A. Studies on the mechanism of action of azinomycin B: Definition of regioselectivity and sequence selectivity of DNA cross-link formation and clarification of the role of the naphthoate. J Am Chem Soc. 2002;124:13008–13017. doi: 10.1021/ja025563k. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, et al. Characterization of a novel DNA glycosylase from S. sahachiroi involved in the reduction and repair of azinomycin B-induced DNA damage. Nucleic Acids Res. 2016;44:187–197. doi: 10.1093/nar/gkv949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Q, et al. Characterization of the azinomycin B biosynthetic gene cluster revealing a different iterative type I polyketide synthase for naphthoate biosynthesis. Chem Biol. 2008;15:693–705. doi: 10.1016/j.chembiol.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Semlow DR, Zhang J, Budzowska M, Drohat AC, Walter JC. Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell. 2016;167:498–511 e414. doi: 10.1016/j.cell.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couvé S, Macé-Aimé G, Rosselli F, Saparbaev MK. The human oxidative DNA glycosylase NEIL1 excises psoralen-induced interstrand DNA cross-links in a three-stranded DNA structure. J Biol Chem. 2009;284:11963–11970. doi: 10.1074/jbc.M900746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couvé-Privat S, Macé G, Rosselli F, Saparbaev MK. Psoralen-induced DNA adducts are substrates for the base excision repair pathway in human cells. Nucleic Acids Res. 2007;35:5672–5682. doi: 10.1093/nar/gkm592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J Biol Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 22.Rubinson EH, Gowda AS, Spratt TE, Gold B, Eichman BF. An unprecedented nucleic acid capture mechanism for excision of DNA damage. Nature. 2010;468:406–411. doi: 10.1038/nature09428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mullins EA, et al. An HPLC-tandem mass spectrometry method for simultaneous detection of alkylated base excision repair products. Methods. 2013;64:59–66. doi: 10.1016/j.ymeth.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullins EA, et al. The DNA glycosylase AlkD uses a non-base-flipping mechanism to excise bulky lesions. Nature. 2015;527:254–258. doi: 10.1038/nature15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parsons ZD, Bland JM, Mullins EA, Eichman BF. A catalytic role for C-H/π interactions in base excision repair by Bacillus cereus DNA glycosylase AlkD. J Am Chem Soc. 2016;138:11485–11488. doi: 10.1021/jacs.6b07399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien PJ, Ellenberger T. The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. J Biol Chem. 2004;279:26876–26884. doi: 10.1074/jbc.M403860200. [DOI] [PubMed] [Google Scholar]
- 27.Brennan RG. The winged-helix DNA-binding motif: Another helix-turn-helix takeoff. Cell. 1993;74:773–776. doi: 10.1016/0092-8674(93)90456-z. [DOI] [PubMed] [Google Scholar]
- 28.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 29.Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 31.Schärer OD, Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. BioEssays. 2001;23:270–281. doi: 10.1002/1521-1878(200103)23:3<270::AID-BIES1037>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 32.Maiti A, Noon MS, MacKerell AD, Jr, Pozharski E, Drohat AC. Lesion processing by a repair enzyme is severely curtailed by residues needed to prevent aberrant activity on undamaged DNA. Proc Natl Acad Sci USA. 2012;109:8091–8096. doi: 10.1073/pnas.1201010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finn RD, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyazono K, et al. A sequence-specific DNA glycosylase mediates restriction-modification in Pyrococcus abyssi. Nat Commun. 2014;5:3178. doi: 10.1038/ncomms4178. [DOI] [PubMed] [Google Scholar]
- 35.Liu M, Imamura K, Averill AM, Wallace SS, Doublié S. Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure. 2013;21:247–256. doi: 10.1016/j.str.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibutani S, Takeshita M, Grollman AP. Translesional synthesis on DNA templates containing a single abasic site: A mechanistic study of the “A rule”. J Biol Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- 37.Yamanaka K, Minko IG, Finkel SE, Goodman MF, Lloyd RS. Role of high-fidelity Escherichia coli DNA polymerase I in replication bypass of a deoxyadenosine DNA-peptide cross-link. J Bacteriol. 2011;193:3815–3821. doi: 10.1128/JB.01550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cundliffe E, Demain AL. Avoidance of suicide in antibiotic-producing microbes. J Ind Microbiol Biotechnol. 2010;37:643–672. doi: 10.1007/s10295-010-0721-x. [DOI] [PubMed] [Google Scholar]
- 39.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 40.Hubbard BK, Walsh CT. Vancomycin assembly: Nature’s way. Angew Chem Int Ed Engl. 2003;42:730–765. doi: 10.1002/anie.200390202. [DOI] [PubMed] [Google Scholar]
- 41.Xu H, et al. Self-resistance to an antitumor antibiotic: A DNA glycosylase triggers the base-excision repair system in yatakemycin biosynthesis. Angew Chem Int Ed Engl. 2012;51:10532–10536. doi: 10.1002/anie.201204109. [DOI] [PubMed] [Google Scholar]
- 42.Igarashi Y, et al. Yatakemycin, a novel antifungal antibiotic produced by Streptomyces sp. TP-A0356. J Antibiot (Tokyo) 2003;56:107–113. doi: 10.7164/antibiotics.56.107. [DOI] [PubMed] [Google Scholar]
- 43.Alseth I, et al. A new protein superfamily includes two novel 3-methyladenine DNA glycosylases from Bacillus cereus, AlkC and AlkD. Mol Microbiol. 2006;59:1602–1609. doi: 10.1111/j.1365-2958.2006.05044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morin A, et al. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]