Table 3.
The posterior predictive analysis of the penguin data for branch length and tree-shape statistics indicating no significant difference in the posterior and posterior predictive tree distributions for model 2 (in Table 2)
| Description | Notation |
 -value -value 
|
|---|---|---|
| Branch length distribution statistics | ||
| The total length of all branches in the tree | 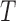 |
0.83 |
| The ratio of the length of the subtree induced by extant taxa and the total tree length |  |
0.33 |
Genealogical Fu and Li’s  calculated as the normalized difference between external branch length in the tree with suppressed sampled ancestor nodes and total tree length. calculated as the normalized difference between external branch length in the tree with suppressed sampled ancestor nodes and total tree length. |
 |
0.5 |
| The time of the MRCA of all taxa |  |
0.57 |
| The time of the MRCA of all extant taxa |  |
0.46 |
| Tree shape statistics | ||
| The maximum number of bifurcation nodes between a bifurcation node and the leaves summed over all bifurcation nodes except for the root |  |
0.75 |
| Coless’s tree imbalance index calculated as the difference between the numbers of leaves on two sides of a node summed over all internal bifurcation nodes and divided by the total number of leaves |  |
0.28 |
| The number of cherries (two terminal nodes forming a monophyletic clade, sampled ancestors are suppressed) | 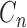 |
0.54 |
| The number of sampled ancestors | SA | 0.26 |
 A
A  value is the proportion of times a given test statistic for the simulated tree exceeds the value of that statistic for the tree from the posterior distribution.
value is the proportion of times a given test statistic for the simulated tree exceeds the value of that statistic for the tree from the posterior distribution.
 All
All  values are within [0.05, 0.95].
values are within [0.05, 0.95].
