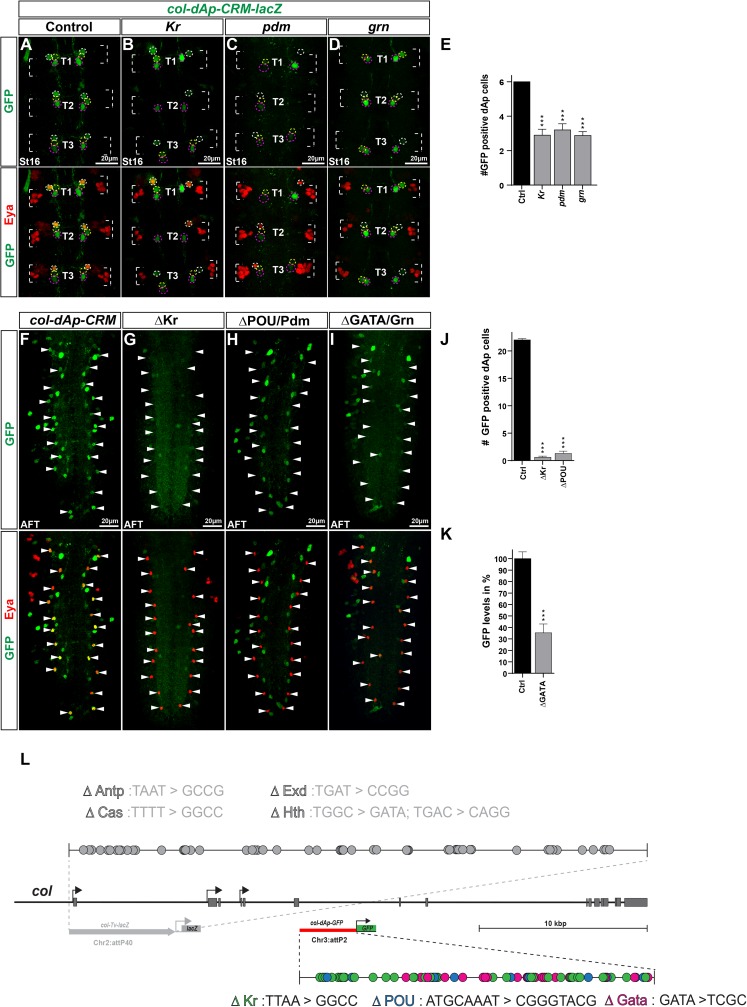
Fig 4. col-dAp enhancer analysis.
(A-D) T1-T3 region of the Drosophila VNC at St16, stained for Eya and GFP to show the activity of the col-dAp-CRM-GFP fragment under (A) control conditions and (B-D) in different mutant backgrounds for col upstream activators in NB4-3 i.e., Kr, pdm and grn. GFP reporter expression under control of the col-dAp-CRM shows reduced expression in all mutant backgrounds. The col-dAp-CRM reports in the dAp cells (white dashed circles), in the sibling cells to the dAp cells (yellow dashed circles) and in one additional cell posterior to dAp sibling cells (magenta dashed circles) are all located in the same plane. (E) Quantification of GFP positive dAp cells in control and mutant backgrounds shows a significant reduction of GFP positive dAp cell numbers in all mutants when compared to control (*** p≤0.0001, n = 10 embryos, Students t-test, +/- SEM). (F-I) Stain for Eya and GFP in whole VNCs at stage AFT to determine the activity of (F) wild type col-dAp-CRM and (G-I) col-dAp-CRMs mutated for potential TFBS for col-dAp upstream activators (indicated by Δ), in the dAp cells. col-dAp-CRMs mutated for potential (G) Kr and (H) Pdm/POU sites, show a loss of GFP signal in the dAp cells. (I) Mutation of potential Grn/GATA binding sites, significantly reduces the GFP signal in the dAp cell. (J) Quantification of GFP positive dAp cells in embryos carrying the col-dAp-CRM-ΔKr or col-dAp-CRM-ΔPOU/Pdm constructs, shows a significant loss of GFP positive dAp cells when compared to control (*** p≤0.0001, n = 10 embryos, Students t-test, +/- SEM). (K) Quantification of GFP levels in dAp cells in flies carrying the col-dAp-CRM-ΔGATA/Grn constructs shows that the levels are significantly reduced when compared to the control (*** p≤0.0001, n≥30 cells, Students t-test, +/- SEM). Schematic representation of the col locus together with the position of the col-dAp-CRM fragment. Color coded dots represent potential transcription factor binding sites (TFBS) found in the col-dAp-CRM fragment, together with the conversion pattern, which was used to mutated the potential binding sites. Chr3:attP3 denotes the landing sites at which all, col-dAp-CRM as well as col-dAp-CRM-ΔTFBS constructs are inserted in the fly genome. Genotypes: (A) col-dAp-CRM. (B) Kr1, KrCD/Kr1, KrCD; col-dAp-CRM/col-dAp-CRM. (C) Df(2L)ED773/ Df(2L)ED773; col-dAp-CRM/col-dAp-CRM (Df(2L)ED773 (removes both nub and Pdm2). (D) col-dAp-CRM/col-dAp-CRM; grn7L12/grnSPJ9. (F) col-dAp-CRM. (G) col-dAp-CRM-ΔKr. col-dAp-CRM-ΔPOU/Pdm. Col-dAp-CRM-ΔGrn/GATA.