Two MYB transcription factors form the core of a gene regulatory network that controls the accumulation and hydroxylation pattern of proanthocyanidins in poplar.
Abstract
The accumulation of proanthocyanidins is regulated by a complex of transcription factors composed of R2R3 MYB, basic helix-loop-helix, and WD40 proteins that activate the promoters of biosynthetic genes. In poplar (genus Populus), MYB134 is known to regulate proanthocyanidin biosynthesis by activating key flavonoid genes. Here, we characterize a second MYB regulator of proanthocyanidins, MYB115. Transgenic poplar overexpressing MYB115 showed a high-proanthocyanidin phenotype and reduced salicinoid accumulation, similar to the effects of MYB134 overexpression. Transcriptomic analysis of MYB115- and MYB134-overexpressing poplar plants identified a set of common up-regulated genes encoding proanthocyanidin biosynthetic enzymes and several novel uncharacterized MYB transcriptional repressors. Transient expression experiments demonstrated the capacity of both MYB134 and MYB115 to activate flavonoid promoters, but only in the presence of a basic helix-loop-helix cofactor. Yeast two-hybrid experiments confirmed the direct interaction of these transcription factors. The unexpected identification of dihydromyricetin in leaf extracts of both MYB115- and MYB134-overexpressing poplar led to the discovery of enhanced flavonoid B-ring hydroxylation and an increased proportion of prodelphinidins in proanthocyanidin of the transgenics. The dramatic hydroxylation phenotype of MYB115 overexpressors is likely due to the up-regulation of both flavonoid 3′,5′-hydroxylases and cytochrome b5. Overall, this work provides new insight into the complexity of the gene regulatory network for proanthocyanidin synthesis in poplar.
Proanthocyanidins (PAs), also known as condensed tannins, are widespread polyphenols with diverse ecological functions. They are polymers of flavan-3-ols and, thus, end products of the phenylpropanoid and flavonoid pathways (Dixon et al., 2005). The PAs are the most broadly distributed secondary metabolites and are especially prominent in forest trees and woody plants (Barbehenn and Constabel, 2011). PA accumulation in trees can be substantial; for example, in some species of poplar (genus Populus), PAs can constitute 25% of leaf dry weight. However, the accumulation of PAs also is highly plastic and varies with genotype and growth conditions (Hwang and Lindroth, 1997; Osier and Lindroth, 2006). In trees, PAs are common constituents of vegetative organs, including roots, leaves, bark, and flowers. Seasonal leaf drop in autumn and turnover of roots thus lead to substantial tannin input into forest soils, where it has been shown to slow litter decomposition and nutrient cycling (Schweitzer et al., 2008). In herbaceous plants, PAs are more restricted in distribution, but they can be found in leaves of legumes, such as birdsfoot trefoil (Lotus corniculatus) and sainfoin (Onobrychis viciifolia; Stringano et al., 2012; Malisch et al., 2015). In berry and tree fruits, PAs and other tannins often are prevalent in unripe fruit, but their concentrations are typically reduced during ripening. In Arabidopsis (Arabidopsis thaliana), PAs can be found only in the seed coat, or testa. The characterization of transparent testa (tt) mutants has led to many key advances in our understanding of the genes required for PA biosynthesis and its regulation (Xu et al., 2015).
PAs are tannins and, thus, functionally defined by their ability to bind and precipitate proteins in solution (Barbehenn and Constabel, 2011). However, in vitro, they also can act as antioxidants and as prooxidants under some conditions (Barbehenn et al., 2006a). These multiple attributes lead to diverse biological functions, in particular in relation to tolerance to environmental stresses. Tannins including PAs have been linked to tree resistance to insect herbivores, but their importance in these interactions has been difficult to establish (Barbehenn and Constabel, 2011). Most tree-feeding insects are lepidopterans, and under the basic midgut conditions of these insects, PAs lose their ability to bind and precipitate proteins via noncovalent interactions. They may cause oxidative stress to the insects, however, and may covalently link to proteins once oxidized (Salminen and Karonen, 2011; Salminen et al., 2011). In vertebrate herbivores, with typically acidic stomachs, tannins do bind dietary protein and demonstrate antinutritive effects when present at high concentrations. Interestingly, in ruminants feeding on protein-rich forages such as alfalfa (Medicago sativa), moderate levels of PAs (less than 6%) are beneficial: by inhibiting microbial activity, PAs prevent rumen foaming and bloating and, thus, reduce the loss of dietary amino acids as urea (Min et al., 2003). PAs also may decrease both methane emissions and nematode burden in ruminants (Novobilský et al., 2013). The presence of PA in unripe fruit is thought to prevent premature feeding prior to seed maturation. Once fruit have ripened, PAs in the epidermis may be important for inhibiting mold due to their broad antimicrobial activity (Scalbert, 1991; Zifkin et al., 2012). In addition, the induction of PA synthesis by biotrophic pathogens may indicate a function in plant defense, although this remains to be demonstrated directly (Miranda et al., 2007). Other proposed roles of PAs in leaves include protection against excess UV light and as free radical scavengers (Close and McArthur, 2002), but the evidence here is limited. Interestingly, many woody plant roots contain substantial concentrations of PAs. Their function here has not been broadly investigated, but PAs have been linked to resistance to Al toxicity in camphor (Cinnamomum camphora) tree roots (Osawa et al., 2011).
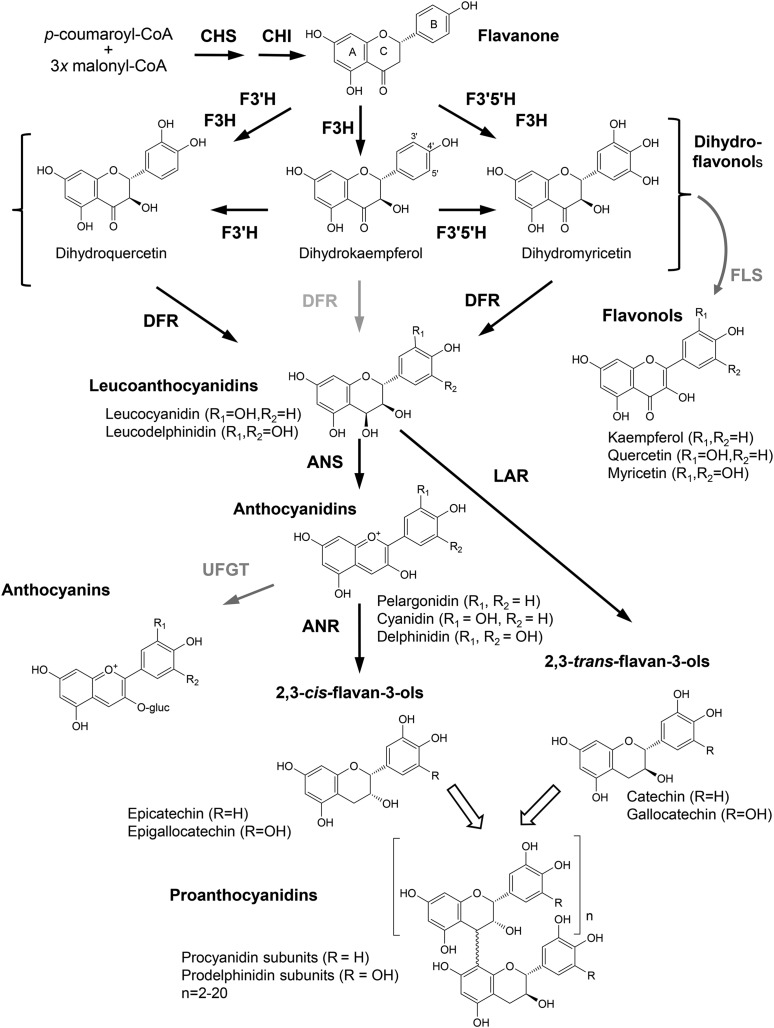
PAs are synthesized via the general flavonoid pathway and share many biosynthetic steps with the anthocyanins (Fig. 1; Dixon et al., 2005). The degree of polymerization of PAs can vary significantly depending on plant species, but it typically ranges from 2 to 20. In addition, the extent of B-ring hydroxylation is variable, with most PAs containing a mixture of dihydroxylated and trihydroxylated B-rings (i.e. procyanidins and prodelphinidins) depending on the species. All major enzymes in the PA pathway are known, and the corresponding genes have been identified. In Arabidopsis, each enzyme is encoded by a single gene, but other species such as poplar may have two or more genes for each biosynthetic step (Tsai et al., 2006). A key discovery in PA biosynthesis was the identification of anthocyanin reductase (ANR), encoded by the BAN gene of Arabidopsis (Xie et al., 2003). This enzyme converts anthocyanidins to epicatechin, the major flavan-3-ol component of PAs. Anthocyanin reductase is specific to PA synthesis and, thus, an important marker for activation of the PA pathway. Anthocyanin-specific UDP-Glc glycosyl transferases can compete with ANR by glycosylating anthocyanidins and diverting them toward anthocyanins (Fig. 1). In the biosynthetic pathway leading to the PAs, the mechanism of polymerization remains a major unresolved question. In Arabidopsis, the MATE transporter TT12 was shown to transport the epicatechin 3-glycoside into the vacuole prior to PA synthesis in the vacuole (Zhao et al., 2010).
Figure 1.
General flavonoid pathway leading to the biosynthesis of PAs. CHS, Chalcone synthase; CHI, chalcone isomerase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, flavonoid 3′,5′-hydroxylase; F3H, flavanone 3-hydroxylase; DFR, dihydroflavonol reductase; FLS, flavonol synthase; ANS, anthocyanidin synthase; LAR, leucoanthocyanidin reductase; ANR, anthocyanidin reductase; UFGT, UDP-Glc flavonoid 3-O-glucosyltransferase. Black arrows indicate the major metabolic routes to PAs in poplar leaves.
Regulation of the PA pathway occurs at the transcriptional level and was first described in Arabidopsis. PA and flavonoid genes in the seed coat were shown to be activated by a complex of three transcription factors: a MYB factor, a basic helix-loop-helix (bHLH) protein, and a WD-repeat (WDR) protein. This MBW complex then binds the appropriate promoters to activate transcription of the biosynthetic pathway genes (Koes et al., 2005; Xu et al., 2015). Specifically, the MYB factor TT2 interacts with the bHLH cofactor TT8 as well as the WDR protein TRANSPARENT TESTA GLABRA1 (TTG1) to specify PA synthesis. TT2 and TT8 proteins both bind specific elements in flavonoid promoters. The WDR does not bind DNA directly but interacts with both TT2 and TT8 and stabilizes the complex (Broun, 2005). TT2 can form functional MBW complexes and induce PA genes by partnering with GLABRA3 (GL3), a distinct bHLH (Koes et al., 2005). Likewise, the bHLH and WDR cofactors can form complexes with other types of MYBs. For example, TT8 interacts with the anthocyanin-specifying MYB PAP1/MYB75 to regulate anthocyanin synthesis. TTG1 also is important in the development of trichomes and root hairs (Ramsay and Glover, 2005). In general, it is the MYB factor that generates specificity: TT2 expression determines PA synthesis, whereas PAP1 specifically regulates anthocyanins. Significant contributions to our knowledge of the MBW complex and its role in anthocyanin synthesis come from petunia (Petunia hybrida) and snapdragon (Antirrhinum majus). For example, petunia AN2 and ROSEA1/2 both belong to the PAP1 subclade of MYBs and have conserved functions in anthocyanin regulation (Koes et al., 2005). Recently, Albert et al. (2014) proposed that the MBW complex contains a dimer of two bHLH proteins.
TT2-type MYB factors have been shown to be key PA regulators in a variety of plant species and organs, including fruit and leaves. In grapevine (Vitis vinifera), persimmon (Diospyros kaki), strawberry (Fragaria × ananassa), and apple (Malus domestica), TT2-like factors and MBW complexes regulate PA synthesis during fruit development (Terrier et al., 2009; Akagi et al., 2010; Schaart et al., 2013; Gesell et al., 2014). Likewise, in leaves and vegetative tissues, PA accumulation is regulated by TT2-like proteins such as LjTT2 in birdsfoot trefoil, TaMYB14 in Trifolium arvense, and PtMYB134 in Populus tremuloides (Yoshida et al., 2008; Mellway et al., 2009; Hancock et al., 2012). These genes all caused enhanced accumulation of PAs when overexpressed. In grapevine, a second type of MYB regulator of PAs, VvMYBPA1, was identified and shown to also activate flavonoid promoters (Bogs et al., 2007). This type of PA regulator has been studied in only a few species, including persimmon and nectarine (Prunus persica; Akagi et al., 2009; Ravaglia et al., 2013). VvMYBPA1 enhances PA accumulation when overexpressed in transgenic grapevine hairy root cultures, but also acts in conjunction with the TT2-like VvMYBPA2 (Terrier et al., 2009). The regulation of PAs is further complicated by the activity of repressor-like R2R3 MYBs, such as VvMYBC2 in grapevine and MYB182 in poplar (Huang et al., 2014; Yoshida et al., 2015). These repressor MYBs interact with bHLH cofactors to inhibit flavonoid gene expression. Anthocyanin regulation is also subject to the action of such MYB repressors (Albert et al., 2014). How the activities of both positive and negative regulators of flavonoids are integrated is a major research question. Furthermore, in trees and woody plants, the transcriptional mechanisms of flavonoid regulation are not extensively studied compared with herbaceous model plants. Trees typically accumulate much greater amounts of phenolics and flavonoids in vegetative tissues, and these may have distinct roles compared with compounds that accumulate in seeds and fruit. Thus, the pattern of regulation of flavonoids in trees may be rather different from that in other plants.
Our long-term aim is to characterize the transcriptional regulation of flavonoid and PA synthesis in Populus. This genus includes poplar, cottonwood, and aspen trees (collectively referred to here as poplars) and is rich in phenolic phytochemicals. The poplar PA pathway can be induced by stresses including herbivory, pathogen attack, UV light, and nitrogen deficiency (Peters and Constabel, 2002; Osier and Lindroth, 2006; Mellway et al., 2009). We first identified the TT2-type stress-inducible PA regulator, MYB134, in Populus tremuloides (Mellway et al., 2009). When overexpressed in transgenic Populus, this MYB causes the hyperaccumulation of PAs in leaves and other vegetative tissues. We also showed that this transcription factor can activate relevant promoters as part of an MBW complex in transiently transformed Arabidopsis leaves (Gesell et al., 2014). This study further characterizes the PA-activating MYBs in Populus. We identified potential downstream targets of MYB134 and identified a second positive regulator, MYB115. Our characterization of this MYB factor, the comparative analysis of both MYB134- and MYB115-overexpressing poplars, and promoter activation assays using both MYBs suggest that these genes act coordinately as part of a complex gene regulatory network. We also discovered that both MYBs, but in particular MYB115, stimulate the flavonoid hydroxylation and prodelphinidin content of PAs, likely via a combination of enhanced flavonoid-3′,5′-hydroxylase and cytochrome b5 expression.
RESULTS
Microarray Analysis of MYB134-Overexpressing Transgenic Poplar Identifies New Poplar MYB PA Regulators
Our previous work demonstrated that MYB134 is an important PA regulator, and transgenic poplars expressing MYB134 show strong increases in the abundance of transcripts encoding the major flavonoid and PA enzymes (Mellway et al., 2009). These plants also accumulated substantially higher concentrations of PAs, but not anthocyanins, flavonols, and other flavonoids; thus, we concluded that the MYB134 gene is a specific regulator of PAs. These plants afforded the opportunity to identify additional PA-related genes, in particular other regulatory genes potentially downstream of MYB134. We took a transcriptomics approach and analyzed differential gene expression in leaves of MYB134 transgenic and control plants using the Affymetrix GeneChip Poplar Whole Genome Array. Using a threshold for differential expression of 2-fold up-regulation (P < 0.05), we identified 167 probe sets representing 110 transcripts with significant overexpression in the MYB134 plants (Supplemental Table S1). Most of the highly up-regulated genes were annotated with functions related to flavonoid synthesis, and one was annotated as a MYB transcription factor. Forty-four genes showed a 5-fold or greater up-regulation in the transgenics, of which 30 encoded flavonoid and phenylpropanoid genes (Supplemental Table S1). No flavonol-, anthocyanin-, or lignin-specific genes were present in this up-regulated gene set, confirming our previous conclusion that the effect of MYB134 is largely restricted to the PA pathway.
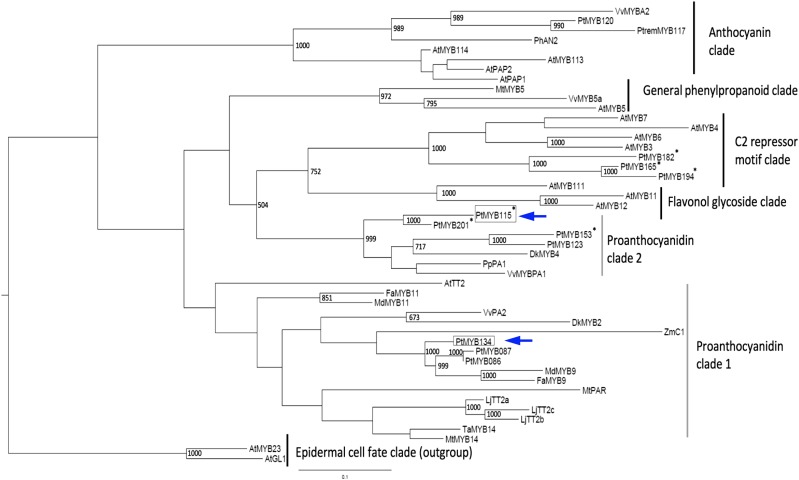
Among the MYB134-up-regulated genes, we identified eight additional MYB transcription factors (Table I). Sequence comparisons indicated that these had similarity to either positive or negative MYB regulators. The positive regulators identified included MYB115, MYB201, and MYB153, based on the naming system used by Wilkins et al. (2009). Of these, MYB115 was the most strongly induced, showing 35-fold enhanced transcript levels (Table I). Phylogenetic analysis determined that MYB115 and MYB201 were very similar, and both clustered with known MYBPA1-type regulators VvMYBPA1 and DkMYB2 (Bogs et al., 2007; Akagi et al., 2010), but separate from MYB134 and TT2-type PA regulators (Fig. 2; Supplemental Fig. S1). Since MYB115 showed the greatest up-regulation and belongs to the MYBPA1 group that has not yet been studied in poplar, we selected this gene for more detailed investigations. Putative negative regulators identified by our transcriptome data included MYB182, MYB165, and MYB194, all with similarity to subgroup 4 R2R3 MYB repressor-like genes, as well as MYB179, a single-repeat R3 MYB repressor-like gene (Table I). We recently demonstrated that MYB182 is a repressor of PAs and anthocyanin genes (Yoshida et al., 2015); the other repressor-like MYBs also appear to have repressor activity (D. Ma and C.P. Constabel, unpublished data).
Table I. MYB transcription factors with elevated transcript levels (greater than 2-fold change; P < 0.05) in MYB134-overexpressing transgenic poplars.
| Probe Seta | Gene Model | Gene Name | Predicted Function | Fold Change | P |
|---|---|---|---|---|---|
| PtpAffx.30659.1.A1_at | Potri.002G173900 | MYB115 | R2R3 MYB, activatorb | 35.30 | 6.23E-12 |
| PtpAffx.8131.6.A1_a_at | Potri.006G221800 | MYB134 | R2R3 MYB, activator | 8.15 | 1.36E-08 |
| PtpAffx.224602.1.S1_at | Potri.008G128500 | MYB194 | R2R3 MYB, repressorc | 7.96 | 1.05E-08 |
| PtpAffx.224650.1.S1_s_at | Potri.010G114000 | MYB165 | R2R3 MYB, repressorc | 3.55 | 7.53E-08 |
| PtpAffx.137746.1.S1_at | Potri.004G088100 | MYB182 | R2R3 MYB, repressorc | 3.53 | 4.79E-08 |
| PtpAffx.224878.1.S1_at | Potri.014G100800 | MYB201 | R2R3 MYB, activator | 3.21 | 1.42E-06 |
| PtpAffx.31264.2.S1_a_at | Potri.015G022000 | MYB179 | R3 MYB, repressord | 2.59 | 2.04E-05 |
| PtpAffx.162546.1.A1_at | Potri.003G144300 | MYB153 | R2R3 MYB, activator | 2.49 | 5.11E-05 |
Three replicate overexpressor and three wild-type plants were probed using the Affymetrix GeneChip Poplar Genome Array.
Sequence similarity to a positive regulator of PA of MYBPA1 type (Bogs et al., 2007).
R2R3 C2 repressor motif.
Single repeat MYB.
Figure 2.
Phylogeny of R2R3 MYB transcription factors involved in PA and flavonoid biosynthesis. Functionally characterized MYB factors, uncharacterized MYBs with enhanced expression in MYB134-overexpressing transgenic poplars, and other poplar and Arabidopsis MYBs grouping with the characterized MYBs are shown. The phylogeny was constructed using the maximum likelihood method, and bootstrap values were calculated from 1,000 phylogenetic constructions. Arrows mark positions of the two MYBs studied here, and asterisks indicate poplar MYB genes showing enhanced expression in MYB134-overexpressing transgenics. Accession numbers are listed in “Materials and Methods.”
We first checked the expression of MYB115 in silico using available online databases (Wilkins et al., 2009), which suggested that MYB115 is expressed in young leaves and roots (Supplemental Fig. S2). These tissues are known to synthesize PAs, and the pattern of expression was similar to that of MYB134 and ANR1, an enzyme specific to the PA branch of flavonoid synthesis. We confirmed the expression pattern of MYB115 in greenhouse-grown P. tremula × P. tremuloides plants by reverse transcription-quantitative PCR (RT-qPCR); this analysis confirmed that leaves and roots showed the highest transcript levels. Developing leaves had the highest MYB115 expression levels of the tissue analyzed (Supplemental Fig. S3). A similar pattern was seen for MYB134 transcripts. To determine if MYB115 also is part of the stress-inducible PA regulatory system similar to MYB134 and flavonoid enzyme-encoding genes, we tested if its expression is enhanced by leaf wounding. Mechanically wounding leaf margins with pliers was used to mimic insect damage. RT-qPCR of RNA from wounded tissue indicated a 4.5-fold up-regulation of the MYB115 transcripts 24 h after wound treatment (Table II). This was comparable to the wound-induced expression of MYB134 (3.3-fold) and further supports a role of MYB115 in PA metabolism.
Table II. Induction of flavonoid and MYB transcripts by leaf wounding in P. tremula × P. tremuloides saplings.
| Transcript | Relative Expression (ΔCt) |
Fold Changeb | Pc | |
|---|---|---|---|---|
| Control | Woundeda | |||
| DFR1 | 0.20 | 1.64 | 8.36 | 0.09 |
| ANR1 | 0.68 | 4.02 | 5.90 | 0.07 |
| MYB134 | 2.55 | 8.40 | 3.29** | 0.02 |
| MYB115 | 0.07 | 0.33 | 4.50** | 0.03 |
Leaf margins were wounded by crushing with pliers 24 h prior to harvest. Leaf plastochron index (LPI) 10 to 12 leaves were collected from each plant and pooled for analysis.
Relative transcript levels in wounded versus control leaves. Boldface values and asterisks indicate significance at the threshold of P ≤ 0.05 (n = 3).
P values indicate results of a one-way ANOVA.
Transgenic Plants Overexpressing MYB115 Show Enhanced Accumulation of PA
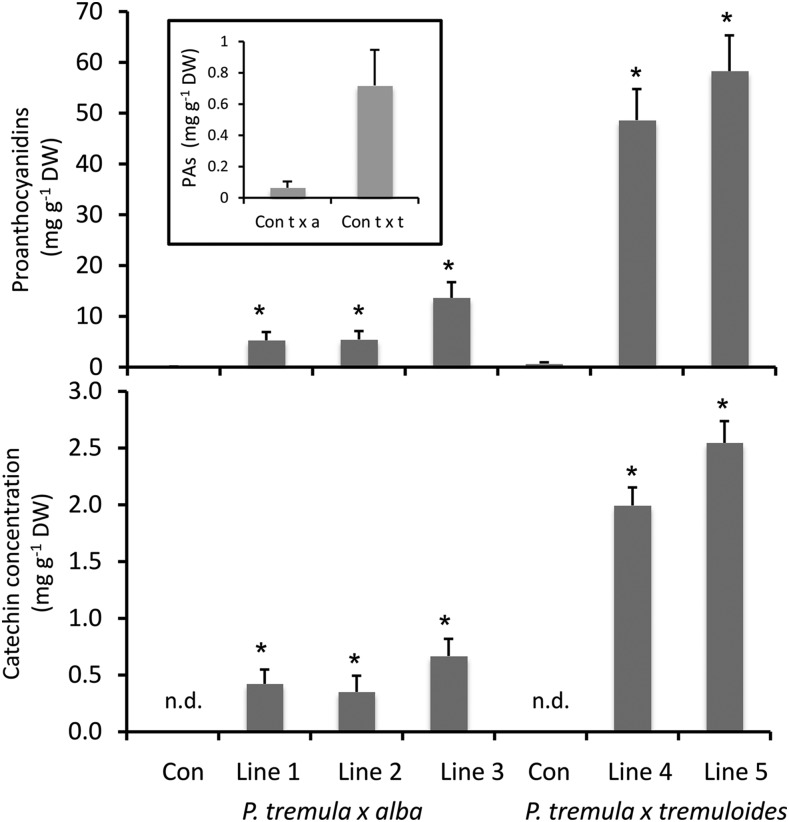
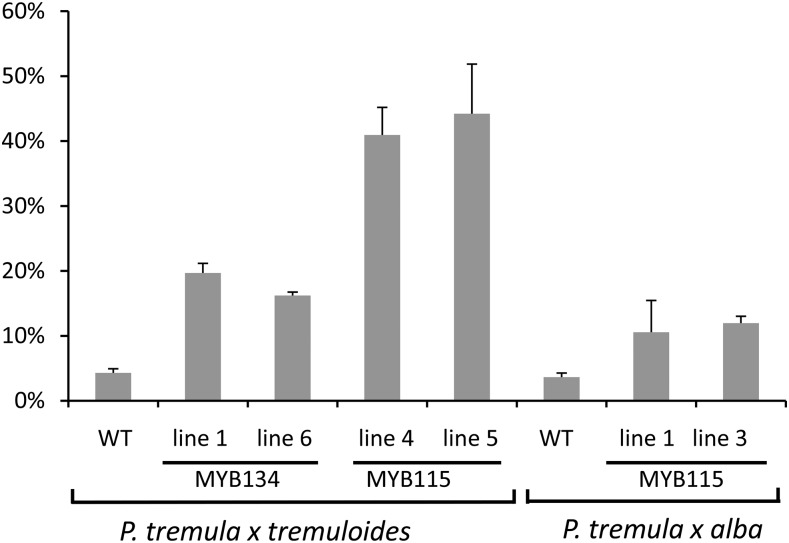
To test the hypothesis that MYB115 is a positive PA regulator, we applied a transgenic approach and generated poplars overexpressing this gene. To facilitate direct comparisons, we used the same vector and cauliflower mosaic virus 35S promoter as for the MYB134 overexpressors. Two poplar hybrid clones were transformed, P. tremula × P. tremuloides (clone INRA 353-38) and P. tremula × P. alba (clone INRA 717-1B4). Putative transformants and MYB115-overexpressing plantlets were identified by PA staining, and several independent high-PA lines were subsequently propagated. Analysis of greenhouse-grown leaves of MYB115-overexpressing lines of both genotypes showed significantly greater PA concentrations compared with controls (Fig. 3). In particular, MYB115 overexpressor lines 4 and 5 (P. tremula × P. tremuloides background) showed more than 50-fold higher PA concentrations than controls. These concentrations are similar to the levels found in the MYB134-overexpressing plants (Mellway et al., 2009); it should be noted that, in the absence of stress, greenhouse plants normally accumulate only very low levels of PAs. Concentrations of catechin, a flavan-3-ol precursor for PAs, showed the same pattern of hyperaccumulation in the MYB115 overexpressors (Fig. 3). Analysis of transcripts by RT-qPCR confirmed the elevated expression of MYB115 in the high-PA lines, as expected (Supplemental Fig. S4). The transgenics showed no morphological abnormalities, and no differences in growth or development were observed.
Figure 3.
Analysis of PAs and catechin in MYB115-overexpressing poplars. PAs in leaf extracts were assayed using the butanol-HCl assay (top), and catechin was measured by HPLC-UV as described (bottom). Transgenic foliage was from two different Populus hybrids, P. tremula × P. alba (clone 717-1B4) and P. tremula × P. tremuloides (clone 353-38). Each bar represents the mean foliar concentration in at least three clonal copies of each independently transformed transgenic line. The inset shows low PA concentrations for both P. tremula × P. alba (t x a) and P. tremula × P. tremuloides (t x t) control (Con) leaves. Error bars indicate se. DW, Dry weight; n.d., not detected. Asterisks indicate significant differences from controls (P < 0.05).
Analysis of Gene Expression in MYB115 Overexpressors and Comparison with MYB134 Overexpression
To obtain a global view of the effects of MYB115 expression and to facilitate direct comparisons with MYB134, we carried out a replicated microarray experiment on MYB115-overexpressing poplar. We used the same microarray platform and experimental design we had used previously for MYB134 analysis. At the differential threshold of 2-fold induction at P < 0.05, we found 208 probe sets to be significantly up-regulated (Supplemental Table S2; Supplemental Fig. S5). In MYB115-overexpressing plants, genes with flavonoid and phenylpropanoid pathway functions dominated the list of strongly induced genes. Among the 59 probe sets that showed a 5-fold or greater differential expression, 29 were annotated for flavonoid or phenylpropanoid pathways, and 12 genes were annotated for transporters and other phenylpropanoid-related enzymes. These results suggest that MYB115, like MYB134, acts as an activator of the PA biosynthesis pathway in poplar.
To better understand the regulation of PA synthesis by these MYB factors, we searched for genes up-regulated in both MYB134- and MYB115-overexpressing plants (Table III). This common set contained genes for all known steps in the flavonoid pathway, including CHS, CHI, F3H, DFR, ANS, F3′H, F3′5′H, ANR, and LAR that were up-regulated in both MYB overexpressors. For each biosynthetic step in the flavonoid pathway, several gene family members were induced in the overexpressors, and at least one gene for each step showed very strong up-regulation (20- to 40-fold). Both general flavonoid and PA-specific biosynthetic genes were affected by MYB134 and MYB115 overexpression.
Table III. Genes related to phenylpropanoid and flavonoid biosynthesis showing increased transcript levels (greater than 2-fold change; P < 0.05) in MYB115-overexpressing (OE) transgenic poplar (line 5) and MYB134-overexpressing transgenic poplar (line 1) as analyzed by Affymetrix GeneChip Poplar Genome Array.
| Probe Set | Gene Model | Gene Name | MYB134 OE |
MYB115 OE |
||
|---|---|---|---|---|---|---|
| Fold Change | P | Fold Change | P | |||
| Flavonoid pathway genes | ||||||
| Ptp.6711.1.S1_s_at | Potri.014G145100 | Chalcone synthase (CHS1) | 6.10 | 1.09E-07 | 33.90 | 3.20E-09 |
| PtpAffx.7896.3.S1_a_at | Potri.001G051600 | Chalcone synthase (CHS3) | 26.52 | 5.64E-09 | 39.65 | 1.80E-11 |
| PtpAffx.7896.2.S1_at | Potri.003G176700 | Chalcone synthase (CHS4) | 41.13 | 1.60E-08 | 64.91 | 9.49E-11 |
| PtpAffx.7896.4.A1_a_at | Potri.003G176900 | Chalcone synthase (CHS6) | 93.77 | 6.18E-09 | 50.99 | 2.07E-11 |
| Ptp.1512.1.S1_s_at | Potri.019G057800 | Chalcone isomerase-like protein (CHIL2) | 4.65 | 1.66E-08 | 7.20 | 5.13E-08 |
| PtpAffx.4850.1.A1_s_at | Potri.010G213000 | Chalcone isomerase (CHI1) | 4.92 | 1.50E-07 | 4.16 | 2.73E-08 |
| Ptp.323.1.S1_s_at | Potri.005G113900 | Flavanone 3-hydroxylase (F3H3) | 14.08 | 8.94E-09 | 2.00 | 3.00E-02 |
| Ptp.4863.1.S1_s_at | Potri.013G073300 | Flavonoid 3′-hydroxylase (F3′H1) | 31.52 | 5.76E-07 | 21.88 | 3.27E-10 |
| PtpAffx.83404.1.A1_at | Potri.009G069100 | Flavonoid 3′,5′-hydroxylase (CYP75A12/F3′5′H1) | 10.20 | 4.24E-08 | 138.39 | 1.45E-07 |
| PtpAffx.37082.1.A1_at | Potri.002G033600 | Dihydroflavonol 4-reductase (DFR1) | 39.54 | 1.32E-08 | 38.51 | 1.21E-09 |
| PtpAffx.25553.1.A1_at | Potri.005G229500 | Dihydroflavonol 4-reductase (DFR2) | 16.57 | 4.01E-04 | 15.61 | 2.33E-08 |
| Ptp.6057.1.S1_at | Potri.001G113100 | Anthocyanidin synthase (ANS2) | 30.75 | 5.77E-11 | 41.49 | 2.15E-09 |
| PtpAffx.5092.1.A1_at | Potri.004G030700 | Anthocyanidin reductase (ANR1) | 28.38 | 1.88E-11 | 20.30 | 6.12E-09 |
| PtpAffx.6065.2.S1_at | Potri.008G116500 | Leucoanthocyanidin reductase (LAR1) | 38.84 | 6.36E-09 | 33.23 | 1.56E-09 |
| Ptp.1080.1.S1_at | Potri.010G129800 | Leucoanthocyanidin reductase (LAR2) | 21.70 | 9.86E-09 | 7.84 | 5.49E-08 |
| PtpAffx.18705.2.A1_a_at | Potri.015G050200 | Leucoanthocyanidin reductase (LAR3) | 60.60 | 1.01E-05 | 21.52 | 2.85E-07 |
| Transcription factors | ||||||
| PtpAffx.224252.1.S1_at | Potri.002G173900 | MYB115 transcription factor | 35.30 | 2.88E-07 | 121.71 | 5.27E-10 |
| PtpAffx.8131.4.A1_a_at | Potri.006G221800 | MYB134 transcription factor | 8.15 | 1.09E-08 | 22.60 | 1.93E-10 |
| PtpAffx.224602.1.S1_at | Potri.008G128500 | MYB194 transcription factor | 7.96 | 3.81E-07 | 2.50 | 4.85E-04 |
| PtpAffx.224650.1.S1_s_at | Potri.010G114000 | MYB165 transcription factor | 3.55 | 2.36E-06 | 3.24 | 7.35E-04 |
| PtpAffx.137746.1.S1_at | Potri.004G088100 | MYB182 transcription factor | 3.53 | 4.17E-08 | 3.74 | 6.71E-05 |
| PtpAffx.224878.1.S1_at | Potri.014G100800 | MYB201 transcription factor | 3.21 | 5.40E-09 | 3.17 | 1.03E-06 |
| PtpAffx.31264.2.S1_a_at | Potri.015G022000 | MYB179 transcription factor | 2.59 | 2.04E-05 | 1.19 | 4.57E-01 |
| PtpAffx.162546.1.A1_at | Potri.003G144300 | MYB153 transcription factor | 2.49 | 5.11E-05 | 1.09 | 5.12E-01 |
| PtpAffx.127289.1.A1_at | Potri.006G209000 | WD40 repeat-containing protein | 5.22 | 5.30E-08 | 16.71 | 2.10E-07 |
| PtpAffx.213439.1.S1_at | Potri.016G075800 | WD40 repeat-containing protein | 4.62 | 7.53E-08 | 2.69 | 2.31E-04 |
| Ptp.8024.1.S1_at | Potri.005G208600 | bHLH131 transcription factor | 3.99 | 6.23E-12 | 3.11 | 7.06E-05 |
| Phenylpropanoid pathway | ||||||
| PtpAffx.161181.1.S1_at | Potri.006G178700 | Cinnamoyl-CoA reductase-like protein | 45.07 | 8.34E-08 | 35.80 | 3.53E-08 |
| PtpAffx.30128.1.S1_at | Potri.001G140700 | Cinnamoyl-CoA reductase-like protein | 8.76 | 2.03E-08 | 6.97 | 1.60E-05 |
| PtpAffx.225544.1.S1_s_at | Potri.016G031000 | p-Coumaroyl shikimate 3′-hydroxylase (C3H3) | 3.33 | 5.48E-09 | 2.56 | 8.08E-06 |
| PtpAffx.225544.1.S1_x_at | Potri.016G031100 | p-Coumaroyl shikimate 3′-hydroxylase (CYP98A23) | 2.62 | 8.21E-07 | 2.38 | 1.49E-05 |
| Ptp.6632.1.S1_at | Potri.019G130700 | Cinnamate 4-hydroxylase (C4H1/CYP73A43) | 2.48 | 2.10E-07 | 8.20 | 3.11E-07 |
| PtpAffx.150025.1.S1_s_at | Potri.013G157900 | Cinnamate 4-hydroxylase (C4H2/CYP73A42) | 2.06 | 6.46E-08 | 7.91 | 8.37E-08 |
| Putative transporters | ||||||
| PtpAffx.224485.1.S1_s_at | Potri.005G207600 | MATE family transporter (AtTT12-like) | 23.98 | 1.05E-08 | 48.94 | 4.81E-10 |
| PtpAffx.94822.1.A1_at | Potri.002G055100 | MATE family transporter (AtTT12-like) | 6.80 | 9.86E-09 | 7.28 | 1.57E-07 |
| PtpAffx.211115.1.S1_at | Potri.013G069200 | MATE family transporter-related | 2.09 | 3.53E-08 | 3.95 | 6.92E-07 |
Similar to the effects of MYB134 overexpression, MYB115 appeared to regulate primarily the PA branch of flavonoid metabolism. Other flavonoid branch pathway genes such as FLS were not affected by MYB115 (Supplemental Tables S1 and S2). Furthermore, only two genes with general phenylpropanoid annotation were up-regulated in both transgenics, cinnamate 4-hydroxylase and p-coumaroyl 3-hydroxylase (Table III). The former is part of the general phenylpropanoid pathway and required to generate p-coumaroyl-CoA (Fig. 1), while the latter catalyzes the subsequent hydroxylation needed for coniferyl and sinapyl alcohol synthesis (Hamberger et al., 2007). Two genes annotated as cinnamoyl-CoA reductase-like showed strong overexpression in both types of transgenics. These have not been characterized but are distinct from the known lignin-specific cinnamoyl-CoA reductases. Both MYB134 and MYB115 transgenics also showed high expression of several TT12-like MATE transporter genes (Table III). Such transporters have been implicated in vacuolar uptake of anthocyanins and PAs in Arabidopsis (Marinova et al., 2007; Zhao et al., 2010).
MYB115 overexpression up-regulated many of the same MYB factors that were originally identified in the MYB134 plants (Table III). MYB134 expression itself was enhanced in MYB115 overexpressors (23-fold), similar to the MYB115 up-regulation in the MYB134 overexpressors (35-fold). This pattern suggests that there is potential for amplification or positive feedback. Except for MYB153 and MYB179, the MYBs we found to be up-regulated in MYB134 plants earlier also were overexpressed in MYB115 plants. Interestingly, the MYB194, MYB165, and MYB182 repressors were contained in this overlapping gene set. Transcripts encoding five other regulatory genes also were up-regulated in both overexpressors: MYB201, MYB153, the bHLH protein bHLH131, as well as two WD40 repeat-containing proteins with similarity to the Arabidopsis TTG1 cofactor. We previously showed that bHLH131 and PtWD40-1 function as cofactors with MYB134 and can activate the ANR1 promoter in transient expression assays (Gesell et al., 2014). Collectively, these data suggest that MYB134 and MYB115 overexpression induced multiple components of a complex transcriptional network that controls PA and flavonoid biosynthesis in poplar. Up-regulation of activators as well as repressors corroborates the idea that this network contains both positive and negative feedbacks (Yoshida et al., 2015).
MYB115 and MYB134 Interact with bHLH131 and Activate Multiple Flavonoid Promoters in Transiently Transformed Poplar Cells
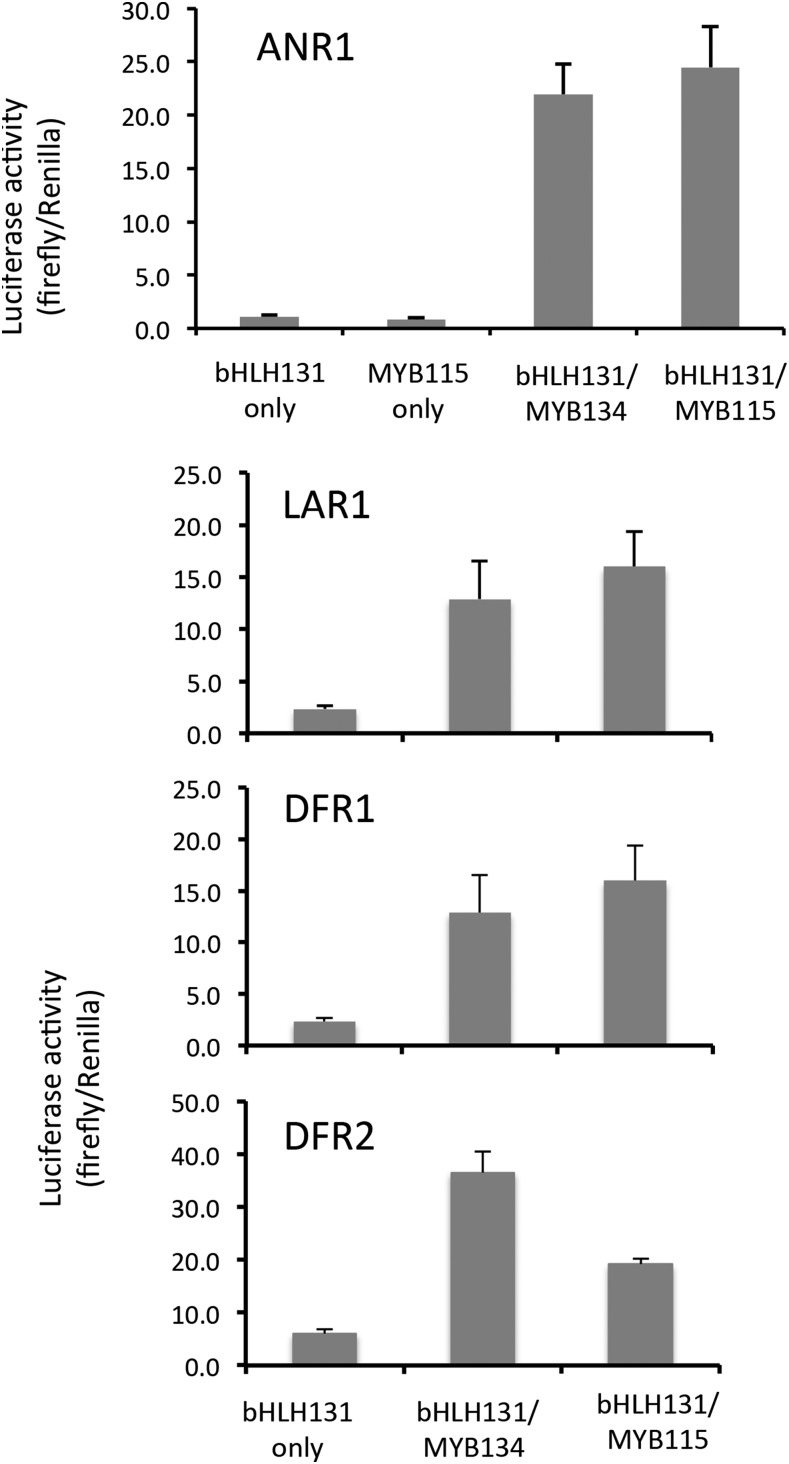
The strongly enhanced transcript levels of many flavonoid and PA genes in both MYB134- and MYB115-overexpressing transgenics suggested that these factors are able to activate multiple flavonoid enzyme promoters. Therefore, we tested if MYB134 and MYB115 could activate other late flavonoid enzyme promoters in transient expression assays. Previously, we had observed that the ANR1 promoter can be activated by MYB134 and that neither the MYB nor the bHLH alone is active (Gesell et al., 2014). We isolated the poplar DFR1, DFR2, and LAR1 promoters, genes that were strongly induced by both MYBs in the transgenics, and constructed vectors for promoter activation assays using the luciferase reporter gene. Effector constructs for MYB115 or MYB134, as well as a construct encoding the bHLH131 cofactor, were transiently expressed in poplar suspension cells. All four promoter constructs were clearly activated by MYB115 and MYB134 when coexpressed with the bHLH cofactor (Fig. 4). No activation was observed in the bHLH-only controls, and MYB115 alone was not active (Fig. 4, top). The similar pattern of activation for each promoter is consistent with the strong overlap of up-regulated genes observed in the transgenics (Table III).
Figure 4.
Activation of flavonoid promoters by MYB134 and MYB115 in transiently transformed poplar suspension cells. Plasmids for promoter-luciferase reporter constructs were cobombarded with effector constructs encoding MYB134, MYB115, and bHLH131 transcription factors as indicated and assayed after a 48-h incubation. Renilla activity is controlled by a constitutive promoter and used to normalize luciferase activity. All data points represent means of at least four biological replicates, and all MYB/bHLH treatments were significantly different from their bHLH-only controls (P < 0.05). Error bars represent se.
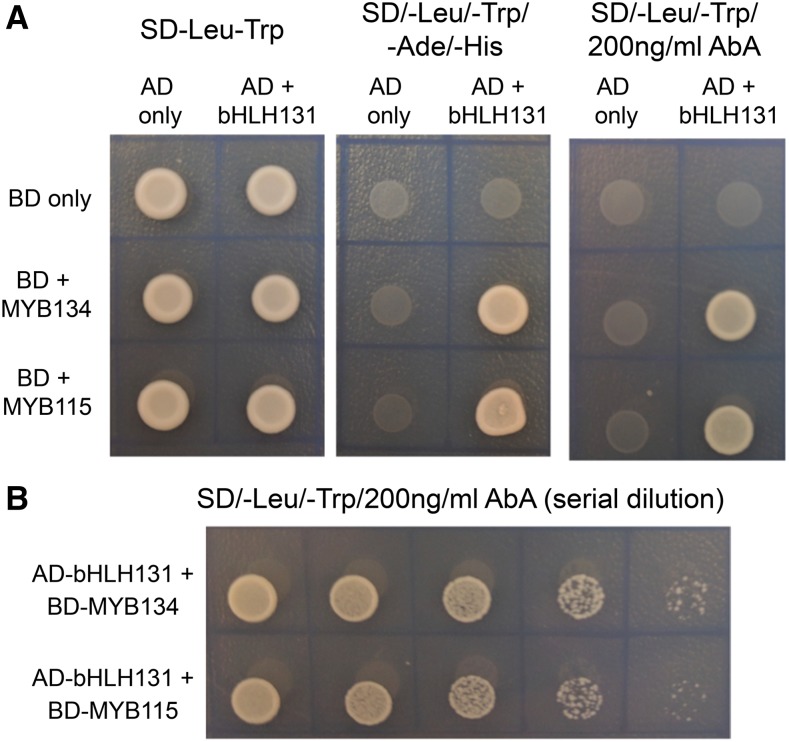
We next tested if the bHLH131 cofactor can interact directly with the MYBs, similar to the Arabidopsis orthologs TT8 and TT2 (Baudry et al., 2006). Both MYB134 and MYB115 contain the six conserved residues within the R3 helix, previously shown to be required for bHLH binding (Supplemental Fig. S1; Zimmerman et al., 2004). Therefore, we carried out yeast two-hybrid experiments to test for this interaction experimentally. Truncated MYB constructs without the C-terminal activation domain were designed, as the presence of this domain led to autoactivation (data not shown). These N-terminal MYB constructs were fused to the binding domain. The bHLH131 constructs included the activation domain. Results with two different yeast marker systems demonstrated that both MYB134 and MYB115 interact directly with bHLH131 (Fig. 5). Dilution experiments using the yeast two-hybrid system indicated that the relative strength of the MYB-bHLH interaction was similar for both MYB134 and MYB115.
Figure 5.
Yeast two-hybrid assay demonstrating the direct interaction of MYB134 and MYB115 with bHLH131. The conserved N-terminal regions of MYB134 and MYB115 were fused to the binding domain (BD) and bHLH131 was fused to the activation domain (AD) of the vector. A, The interaction was tested in the yeast two-hybrid assay using two selectable markers, the His/Ade auxotrophs (middle panel) and Aureobasidin A (AbA; right panel). Control colonies with no selection are shown on the left panel. When fused to the BD vector, MYB134, MYB115, and bHLH131 all have autoactivation (data not shown). B, Serial dilution (5-fold dilutions) of the two-hybrid assay with AbA.
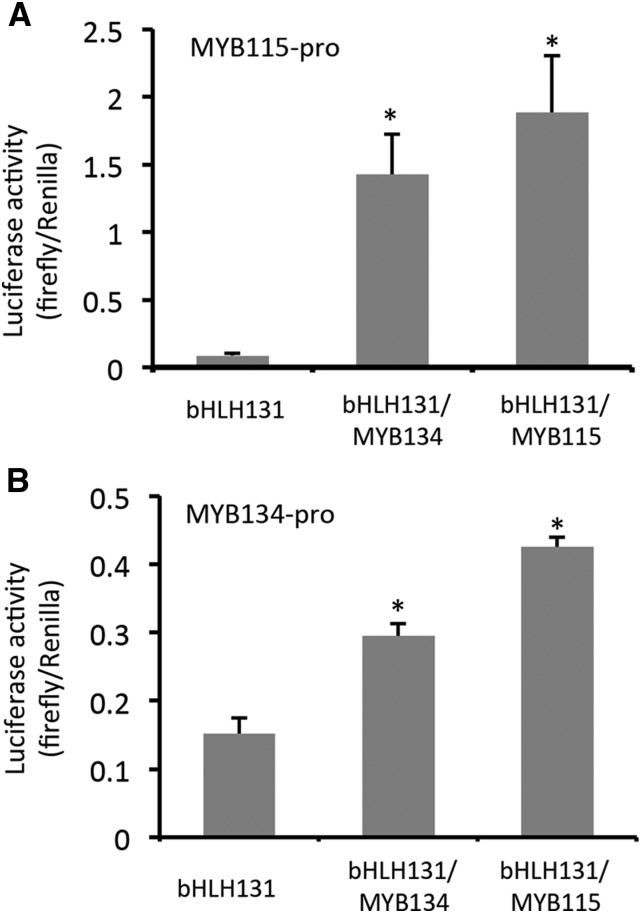
The induction of MYB115 transcripts in MYB134-overexpressing plants as well as the reciprocal induction of MYB134 in MYB115 overexpressors suggested that these MYBs might be subject to regulation by each other. We thus isolated the MYB115 and MYB134 promoters and tested these in our transient expression assay in poplar suspension cells. In combination with bHLH131, MYB134 clearly activated the MYB115 promoter (Fig. 6A). Surprisingly, when MYB115 itself was expressed together with the MYB115 promoter-luciferase construct, we found that MYB115 could activate its own promoter. In addition, when we tested a MYB134 promoter-luciferase construct, we observed a similar result: both MYB115 and MYB134 moderately activated the MYB134 promoter (Fig. 6B). Therefore, both of these transcription factors appear to activate each other’s, as well as their own, promoters, which could provide potential positive feedback loops.
Figure 6.
Activation of the MYB115 and MYB134 promoters by MYB134 and MYB115 in transiently transformed poplar cells. MYB115 promoter (A) or MYB134 promoter (B) constructs were introduced into poplar cells together with expression constructs for bHLH131, MYB115, or MYB134, as described in “Materials and Methods.” Renilla is controlled by a constitutive promoter and used to normalize luciferase activity for each transformation. Asterisks indicate luciferase activity significantly different from the bHLH-only controls (Student’s t test, P < 0.05). Error bars indicate se.
MYB115 Overexpression Leads to Reduced Concentrations of Salicinoid Phenolic Glycosides and Enhanced Flavonoid B-Ring Hydroxylation
To test for potential effects of MYB115 overexpression on other soluble phenolics, we compared these in leaves of MYB115 overexpressor and control plants using HPLC. In both the P. tremula × P. alba and P. tremula × P. tremuloides genetic backgrounds, MYB115 overexpression led to a clearly reduced concentration of salicortin and tremulacin, two antiherbivore phenolic glycosides derived from salicyl alcohol (Table IV; Boeckler et al., 2011). The transgenic lines with the greatest enhancement of PAs, lines 4 and 5 in the P. tremula × P. tremuloides background, also showed the largest reduction in salicinoids (Table IV; Fig. 3). Salicin was much less abundant than salicortin or tremulacin and was only reduced by MYB115 overexpression in the P. tremula × P. alba background. The overall pattern is comparable to that of the MYB134 overexpressors, where we had also reported strongly reduced levels of tremulacin and salicortin (Mellway et al., 2009; Boeckler et al., 2014).
Table IV. Concentrations of major salicinoids and dihydromyricetin in MYB115-overexpressing and control leaves as determined by HPLC-UV.
Concentrations are expressed as mg g−1 dry weight ± se. Significant differences from controls were determined by one-way ANOVA and are indicated by boldface type and asterisks (*, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001). n.d., not detected.
| Compound |
P. tremula × P. tremuloides (353-38) |
P. tremula × P. alba (717-1B4) |
|||||
|---|---|---|---|---|---|---|---|
| Control | MYB115 Line 4 | MYB115 Line 5 | Control | MYB115 Line 1 | MYB115 Line 2 | MYB115 Line 3 | |
| Salicortin | 90.05 ± 20.07 | 15.18 ± 0.79* | 17.76 ± 3.19* | 41.92 ± 3.04 | 21.29 ± 3.52** | 19.87 ± 7.54* | 16.22 ± 4.78** |
| Tremulacin | 76.26 ± 13.34 | 12.71 ± 2.15** | 14.37 ± 3.43** | 60.61 ± 7.60 | 31.56 ± 7.59* | 29.61 ± 12.12 | 22.88 ± 7.32** |
| Salicin | 0.48 ± 0.13 | 0.33 ± 0.01 | 0.33 ± 0.06 | 3.19 ± 0.10 | 0.51 ± 0.07*** | 0.35 ± 0.13*** | 0.43 ± 0.10*** |
| Dihydromyricetin | n.d. | 3.00 ± 0.65 | 4.68 ± 1.87 | n.d. | n.d. | n.d. | n.d. |
Further comparison of the phenolic profiles led to the identification of other peaks with altered abundance in the high-PA MYB115 overexpressors. In particular, a prominent peak was observed in HPLC profiles of MYB115 overexpressors in the P. tremula × P. tremuloides background but not in controls (Supplemental Fig. S6). This peak was not present in either transgenic or control leaves of the P. tremula × P. alba genotype. Liquid chromatography-mass spectrometry (LC-MS) analysis of the peak gave a mass-to-charge ratio (m/z) value of 321 in positive mode and a prominent fragment at m/z 275. The compound also ionized in negative mode to show a signal at m/z 319 with a fragment in tandem mass spectrometry (MS/MS) at m/z 193. Based on its mass and fragments, and ultimately by comparison of the mass spectra and the retention time with those of the commercially available standard, we identified this peak as dihydromyricetin, a dihydroflavonol hydroxylated at the 3′, 4′, and 5′ positions of the B-ring. This compound had been observed as a minor component in bark and leaf extracts of Populus (Pearl and Darling, 1969). Although we did not identify dihydromyricetin in our earlier phytochemical analysis of MYB134 plants (Mellway et al., 2009), reanalysis of those plants determined that this compound was indeed present, again only in the P. tremula × P. tremuloides background (data not shown). Dihydromyricetin is a predicted intermediate in the synthesis of gallocatechin and epigallocatechin, two of the flavan-3-ols that are presumed precursors for PAs (Fig. 1; Scioneaux et al., 2011). Dihydromyricetin also is an intermediate for the biosynthesis of other flavonoids with 3′, 4′, and 5′ hydroxylation on the B-ring, in particular the flavonol myricetin and the anthocyanidin delphinidin. Myricetin glycosides were reported previously for Populus balsamifera (Pearl and Darling, 1971). Based on HPLC-UV quantification using the authentic standard, we determined that the content of dihydromyricetin in the MYB115 transgenics was 3 to 5 mg g−1 leaf dry weight (Table IV).
The accumulation of dihydromyricetin in the MYB115 overexpressors suggested that other flavonoid end products, including the PAs, could also show greater B-ring hydroxylation. Poplar PAs contain a mixture of procyanidin (dihydroxylated B-ring) and prodelphinidin (trihydroxylated B-ring) subunits. The ratio of these monomers varies in Populus species (Ayres et al., 1997; Scioneaux et al., 2011), but how this is regulated is not known. Catechin and gallocatechin, flavan-3-ol precursors of procyanidins and prodelphinidins, respectively, were detected in both MYB overexpressors using LC-MS; in control plants, concentrations were too low to be reliably quantified. However, in MYB115 overexpressors in the P. tremula × P. tremuloides background, we were able to estimate a gallocatechin:catechin ratio of approximately 1:3, based on the total ion current in LC-MS peaks. This suggested that the PAs in our transgenics also could be high in trihydroxylated B-rings (i.e. prodelphinidin subunits). To directly test this idea, a subset of the MYB overexpressor and control leaf extracts was subjected to more detailed analysis using a novel PA-fingerprinting and quantification method (Engström et al., 2014). This ultra-performance liquid chromatography (UPLC)-MS/MS method is based on the in-source fragmentation of PA oligomers and polymers, using a series of different cone voltages for each sample. Each voltage thus gives rise to optimal fingerprints depending on polymer size, and these can be quantified using multiple reaction monitoring (MRM) for prodelphinidin and procyanidin subunits. We previously showed that procyanidin and prodelphinidins from a range of polymer sizes could be quantified accurately with this method (Engström et al., 2014). This approach is ideal for determining the B-ring hydroxylation of transgenic poplar PAs, since it quantifies procyanidin and prodelphinidin subunits for the polymer individually.
The fingerprints obtained show clear differentiation of subunits consistent with a shift in B-ring hydroxylation (Supplemental Fig. S7). Based on these MRM chromatograms and standard curves, we determined prodelphidin and procyanidin contents and calculated the percentage of prodelphinidin for the PAs (Fig. 7). We observed a high percentage of prodelphinidin in both MYB transgenic types, with a proportion of prodelphinidin of approximately 40% in MYB115 and approaching 20% in MYB134 overexpressors in the P. tremula × P. tremuloides background. By contrast, the MRM chromatograms revealed that control leaves had proportionally less prodelphinidins relative to procyanidins than the MYB overexpressing leaves (Supplemental Fig. S6, bottom). While in controls, procyanidin and prodelphinidin fingerprint signals were substantially weaker than in the MYB transgenics, they were above the limit of detection and gave a prodelphinidin proportion of approximately 4% (Engström et al., 2014; Supplemental Table S3). In the P. tremula × P. alba background, we also detected a clear increase in the proportion of prodelphinidins, from 4% in controls to 10% to 12% in MYB115 overexpressors (Fig. 7; Supplemental Table S3). We also analyzed the most abundant flavonols, since these compounds are derived directly from dihydroflavonols (Fig. 1). Whereas control plants accumulated primarily kaempferol and quercetin glycosides, transgenics overexpressing either MYB additionally accumulated myricetin glycosides (Supplemental Table S4). This effect was again particularly strong in the MYB115 plants.
Figure 7.
Mean percentage of prodelphinidins in total PA in MYB134- and MYB115-overexpressing transgenic Populus plants. The composition of PA was quantified by UPLC-MS/MS as described in “Materials and Methods.” Data for multiple independent transgenic lines in both hybrid backgrounds (P. tremula × P. tremuloides and P. tremula × P. alba) are shown. All transgenic lines are significantly different from their respective wild-type (WT) controls (P < 0.05, Student’s t test), with P. tremula × P. alba MYB115 line 1 approaching significance (P = 0.055). Error bars indicate se; n = 3 or 4 for all transgenic lines.
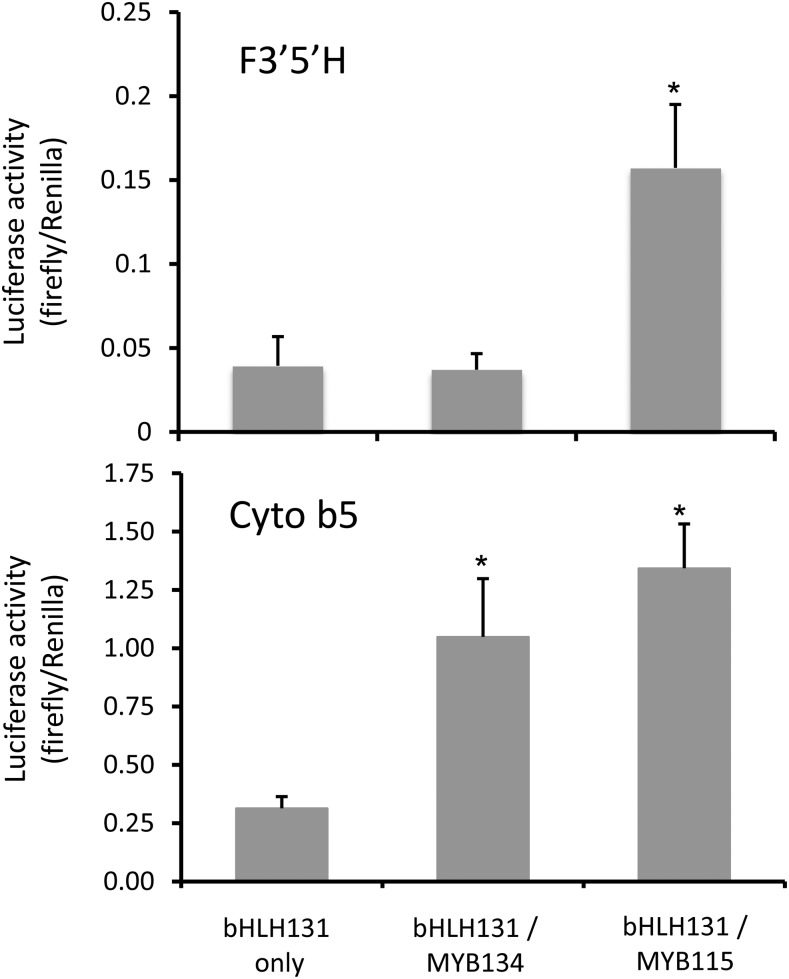
Flavonoid hydroxylation patterns are due to the relative activities of F3′H and F3′5′H, enzymes that act early in the flavonoid pathway and, thus, affect the structure of several end products (Fig. 1). We inspected our transcriptome data and found that F3′5′H transcripts were 138-fold more abundant in MYB115 overexpressors compared with controls, much more than F3′H (22-fold; Table III; Supplemental Table S2). F3′5′H was the most strongly up-regulated transcript in these plants, suggesting that this gene could be directly responsible for the strong hydroxylation shift. To directly test if F3′5′H is activated by MYB115, we used F3′5′H promoter-luciferase constructs in our transient activation assays. MYB115 clearly activated the F3′5′H promoter (Fig. 8), matching the strong enrichment of F3′5′H transcripts and the hydroxylation shift in MYB115 overexpressor plants. By contrast, MYB134 was not active with the F3′5′H promoter. This result suggested that the observed hydroxylation shift in MYB134 overexpressors may not be directly regulated via transcription of this enzyme; furthermore, up-regulation of F3′5′H transcripts was only 10-fold in these plants, significantly less than the induction of F3′H (Table III).
Figure 8.
Activation of F3′5′H and cytochrome b5 promoters in transiently transformed poplar suspension cells. The F3′5′H and cytochrome b5 promoter fragments were cloned upstream of the luciferase reporter gene. Renilla was used as a normalizer gene. Plasmids for promoter-luciferase reporter constructs were cobombarded with MYB and bHLH effector constructs and assayed as described. All data points represent means of at least four biological replicates. Error bars represent se.
To look for additional potential controls of PA and flavonoid hydroxylation, we further investigated the MYB plant transcriptome data. Significantly, we observed a very strong up-regulation of the cytochrome b5 gene in both MYB134 and MYB115 overexpressors, with 80- and 165-fold up-regulation, respectively (Supplemental Tables S1 and S2). Cytochrome b5 is uniquely required as a second reductase for F3′5′H activity, but not for F3′H or most other P450 enzymes (de Vetten et al., 1999; Renault et al., 2014). Therefore, we cloned and assayed the cytochrome b5 promoter in the transient activation assay as above; this promoter was activated by both MYB134 and MYB115 (Fig. 8). This result could indicate that cytochrome b5 is a key player in the regulation of flavonoid hydroxylation in poplar, as its activation by both MYBs is consistent with the hydroxylation shift and its enhanced expression in both transgenics.
DISCUSSION
The importance of MYB factors in the regulation of PA biosynthesis in seeds and fruit has been demonstrated in several species, but few studies have investigated their role in plants that accumulate PAs in leaves. Here, we report on further molecular characterization of the poplar MYB134 transcription factor and the discovery of MYB115, a second stress-responsive MYB regulator that belongs to the MYBPA1 class. Both genes are specific activators of PA metabolism in poplar, and their activities in transgenic plants help to identify additional MYB regulators linked to this pathway. These genes provide the outlines of a gene regulatory network for PAs in poplar.
The Poplar MYB115 Gene Encodes a New MYBPA1-Type Regulator Active in Leaves
MYB115 belongs to a subclade of flavonoids regulating MYBs that was defined by the MYBPA1 gene in grapevine and is distinct from the well-studied TT2 group first discovered in Arabidopsis. Although MYB115 orthologs have been studied in fruit of grapevine, persimmon, nectarine, and cocoa (Theobroma cacao; Terrier et al., 2009; Akagi et al., 2010; Ravaglia et al., 2013; Liu et al., 2015), our study focuses on a member of this group that is active in leaves. Our data showed that MYB115 transcripts were induced by wounding, and gene expression profiling confirmed that MYB115 is highly expressed in developing leaves where the PA pathway is most active. Overexpressing MYB115 led to the strong up-regulation of all known steps of PA synthesis but did not induce flavonol synthase or lignin-specific genes. Likewise, phytochemical profiling demonstrated an enhanced accumulation of PAs but did not have major effects on anthocyanins or flavonols. These data confirm that MYB115 primarily affects PA metabolism. This conclusion is supported by promoter activation assays in transiently transformed poplar cells, which demonstrate that MYB115 can activate relevant flavonoid promoters.
Our cumulative data suggest that MYB134 and MYB115 have similar effects on flavonoid and PA gene expression. Both activate a similar set of promoters in transiently transformed poplar cells, and the comparative transcriptomic analysis of MYB134 and MYB115 overexpressor plants demonstrates a high degree of overlap. Likewise, the phenolic profiles of these plants were similar: high accumulation of PAs, elevated concentrations of the PA precursor catechin, greater hydroxylation of PA, and reduced accumulation of salicinoid phenolic glycosides (Table III; Mellway et al., 2009). MYB115 does appear to have a stronger effect on F3′5′H expression, more strongly impacting the prodelphinidin content of the PAs compared with MYB134 (see below). Some of these effects are reminiscent of prior work on the grape orthologs of MYB115 and MYB134, VvMYPA1 and VvMYBPA2 (Terrier et al., 2009). When overexpressed in hairy roots, the grape MYBs also induced a largely overlapping set of flavonoid genes and were specific to PAs. Likewise, Akagi et al. (2009, 2010) reported both TT2- and MYBPA1-type regulators for persimmon, named DkMYB2 and DkMY4, respectively, and demonstrated that both interact directly with flavonoid and PA-associated gene promoters. By contrast, in Arabidopsis, only TT2-type PA MYB regulators are known; this could reflect the restriction of PA synthesis to the developing seed coat.
MYB115 and MYB134 Are Components of a Gene Regulatory Network for PA Synthesis
Since we first identified MYB115 based on its transcript abundance in MYB134-overexpressing poplars, we hypothesized that it might itself be regulated by MYB134. We corroborated this idea using transient expression experiments, which showed that MYB134 activated the MYB115 promoter. However, in the converse experiment, MYB115 also activated MYB134 (Fig. 6). This suggests that both MYBs have redundant and dual functions: in addition to activating the flavonoid pathway directly, they can induce the expression of additional PA regulatory genes similar to feed-forward loops that can act as persistence indicators (Mangan and Alon, 2003). It is interesting that in both MYB134- and MYB115-overexpressing stable transgenics, transcripts of the other MYB also were up-regulated, consistent with the transient promoter activation data. Therefore, our data are most consistent with parallel, or redundant, functions of both MYBs in the regulatory network. RNA interference plants with suppressed MYB134 expression are in progress and should be able to help determine to what extent the functions of MYB115 and MYB134 overlap.
Furthermore, our transient transformation experiments demonstrated that both MYB115 and MYB134 activated their own and each other’s promoters (Fig. 6). This suggests additional mechanisms for positive feedback in PA induction. Such positive feedback loops could serve to amplify an induction signal in response to stress. Autoregulation of transcriptional activators involved in the flavonoid biosynthetic pathway has been reported previously. For example, the Arabidopsis TT8 gene was shown to regulate its own promoter during PA accumulation in the seed coat (Baudry et al., 2006). Likewise, in red-fleshed cultivars of apple, the MdMYB10 gene is subject to autoregulation due to tandem repeats that form a minisatellite-like structure in the MdMYB10 promoter (Espley et al., 2009). In addition, in our poplar microarray data, both MYB134 and MYB115 overexpression stimulated elevated transcript levels of MBW cofactors, bHLH131 and the WD40 protein. We previously showed that a bHLH cofactor is required in order for MYB134 and MYB115 to function in the transient promoter activation assay in vivo; the induction of MBW cofactors by both activator MYBs could act as an additional positive feedback loop leading to amplification of the signal.
Our transcriptome data also suggest the existence of negative feedback in the regulation of PA biosynthesis. Both MYB overexpressor plants had significantly enhanced expression of the MYB182, MYB165, and MYB194 genes, all encoding MYB repressors. We had demonstrated previously in transient expression assays using the ANR1 promoter that MYB182 represses the MYB activators and also reduces the accumulation of PAs and anthocyanins when overexpressed in transgenic plants (Yoshida et al., 2015). Subsequent experiments have shown that MYB182, MYB165, MYB194, as well as MYB179, are active as repressors in transient expression assays with several flavonoid promoters, including ANR1, DFR1, and ANS (D. Ma and C.P. Constabel, unpublished data). Thus, the new MYB repressors uncovered by our array experiments also could represent potential nodes in the PA gene regulatory network. The physiological functions of these negative regulators are not yet fully known, but hypothetical roles include preventing the overaccumulation of PAs and down-regulating competing biosynthetic pathways. The latter function is consistent with the observation that, despite the up-regulation of many general flavonoid pathway genes, only the PAs accumulate to a substantive extent. Similarly, the reduced salicinoid content we observed in both MYB overexpressors suggests that some repressor MYBs may act to reduce salicinoid biosynthesis (see below).
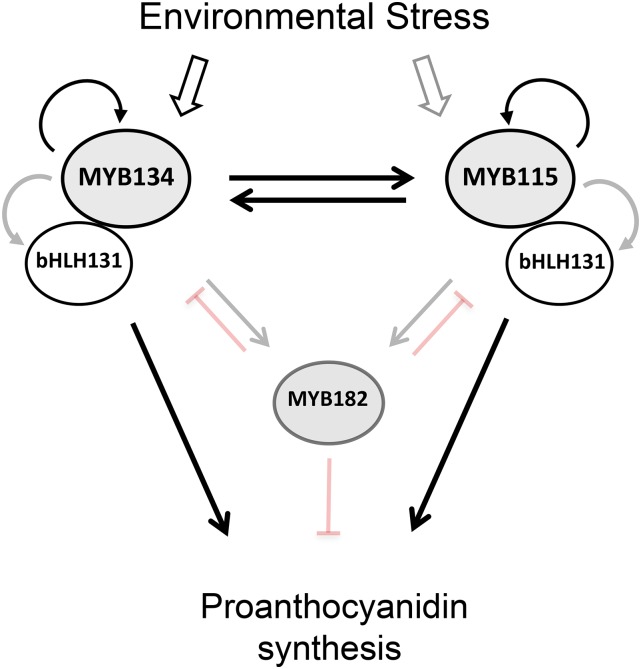
The observed interactions are summarized in a tentative model of the PA regulatory gene network (Fig. 9). Based on our promoter activation and transcriptomics data, MYB134 and MYB115 activate each other as well as the bHLH131 and WD40 cofactors. Both MYB134 and MYB115 regulate flavonoid biosynthetic genes. The MYB182 repressor is downstream of both MYB activators but, in turn, dampens the induction of both flavonoid enzyme genes and several regulatory genes, including the bHLH. This multilevel activity is reminiscent of the double lock-down model described for the regulation of anthocyanin synthesis in petunia (Albert et al., 2014). In that species, activator and repressor MYBs were shown to modulate the expression of genes encoding both flavonoid enzymes and the components of the MBW complex itself. Some elements of our model are similar to those suggested for PA synthesis in grapevine, where both positive and negative MYB regulators are known, including R2R3 MYB factor repressors that restrict PA synthesis at the level of flavonoid enzymes and the MBW genes (Terrier et al., 2009; Huang et al., 2014; Cavallini et al., 2015). Our study integrates a broader diversity of data, in particular the direct transcriptional activation assays using relevant promoters together with transcriptomic analysis of transgenic poplar plants. In this regard, poplar is particularly useful as an experimental system. With the ongoing characterization of additional MYB repressors, their promoters, and targets, it will be possible to continue to elaborate on this model.
Figure 9.
Schematic summarizing positive and negative interactions among transcription factors known to regulate PA synthesis in poplar. Heavy black lines indicate activation demonstrated both via transient expression assays and in transgenic MYB-overexpressing plants, while gray lines indicate the up-regulation observed in transgenic overexpressor plants only. Light black lines indicate interactions shown by transient activation only. Inhibitory interactions with MYB182 are from Yoshida et al. (2015).
MYB134 and MYB115 Overexpression Leads to Reduced Accumulation of Phenolic Glycosides and a Greater Extent of B-Ring Hydroxylation in PAs and Flavonoids
The overexpression of MYB134 and MYB115 regulators and subsequent overaccumulation of PAs led to unexpected alterations in other phenolic pathways. We had previously observed that the MYB134 overexpressors accumulated 2- to 3-fold lower concentrations of the major poplar salicinoids, salicortin and tremulacin (Mellway et al., 2009). Here, we demonstrate a similar reduction in salicinoids in MYB115-overexpressing plants. Thus, high flux into flavonoid and PA biosynthesis is associated with reduced salicinoid synthesis in both MYB134- and MYB115-overexpressing transgenics, suggesting a direct interconnectedness of these phenolic pathways in poplar. Indirect effects on soluble phenolics were observed previously in transgenic poplar with suppressed expression of p-coumaroyl-CoA 3′-hydroxylase, a key enzyme of monolignol metabolism (Coleman et al., 2008). It remains to be determined whether the MYB134 and MYB115 transgenes can cause a direct repression of these phenolic pathways at the transcriptional level, for example by increasing the abundance or activity of transcriptional repressors. The discovery of the new MYB repressor genes induced by MYB134 and MYB115 provides a potential mechanism for the down-regulation of competing pathways. Although salicinoid biosynthesis is not yet elucidated and no enzymes have been demonstrated conclusively, candidate genes have been identified (Chedgy et al., 2015) and their promoters could be potential targets for the repressors. Tradeoffs in allocation to either PAs or salicinoids have been reported in P. tremuloides and are thought to reflect distinct adaptive strategies of different genotypes (Holton et al., 2003). Identifying the molecular mechanism underlying such adaptive strategies is a long-term research objective of our research in Populus.
Our analysis demonstrates that MYB overexpression enhanced the proportion of prodelphinidin, reaching almost 50% of total PAs in the MYB115 overexpressors. The shift toward greater trihydroxylation of the flavonoid B-ring ultimately depends on increased F3′5′H activity; however, while the F3′5′H gene was highly up-regulated in MYB115 transgenics, this was much less pronounced in MYB134 plants. Unexpectedly, we also observed that the cytochrome b5 gene, known to be a second electron donor for F3′5′H but not most other cytochromes P450, was up-regulated strongly in both MYB115 and MYB134 overexpressors. Correspondingly, in transient promoter activation assays, the cytochrome b5 promoter was activated by both MYBs. However, only the F3′5′H promoter responded to MYB115, matching the observed transcriptome patterns in the MYB-overexpressing transgenics. These data, together with the greater B-ring hydroxylation of PAs in both types of transgenics, suggest that cytochrome b5 could be a limiting factor and determine F3′5′H activity and B-ring hydroxylation in poplar. This could explain why the proportion of prodelphinidin is particularly elevated in MYB115 plants, since these transgenics expressed both the F3′5′H and cytochrome b5 genes to high levels.
Cytochrome P450 monooxygenases require two electrons, which are typically provided by P450 oxygenase reductase (Renault et al., 2014). In the case of F3′5′H activity, however, one of the two electrons is provided by cytochrome b5. This cooperation of cytochrome b5 with F3′5′H was first discovered in petunia, where the difF mutation resulted in flowers that lack delphinidin. This phenotype was subsequently found to be due to a nonfunctional cytochrome b5 (de Vetten et al., 1999). Mutations in this gene resulted only in a loss of F3′5′H-dependent flower color. F3′H activity does not require cytochrome b5; thus, cyanidin derivatives were not affected. In other plants and tissues that synthesize delphinidin derivatives, such as ripening grape berries and blueberry (Vaccinium corymbosum) fruit, cytochrome b5 also is tightly coexpressed with anthocyanin genes (Bogs et al., 2007; Zifkin et al., 2012). Cytochrome b5 is needed in some species, together with F3′5′H, for the genetic engineering of delphinidin synthesis and purple flower color (Tanaka and Brugliera, 2013). It is not known why F3′5′H, unlike most other P450 monooxygenases in plants, requires cytochrome b5. However, our observations suggest that cytochrome b5 regulation is a potential mechanism for determining flavonoid hydroxylation in poplar.
At a higher level, the relative activity of MYB115 and MYB134 in poplars could determine the degree of B-ring hydroxylation in PAs via the differential regulation of F3′5′H and cytochrome b5 by these transcription factors. Controlling the degree of PA hydroxylation could represent an important adaptation to fine-tune the response to stress. The extent of B-ring hydroxylation of PAs is important for the biological activity of the PAs; for example, PAs with a larger proportion of prodelphinidin were correlated with greater antiherbivore activity (Ayres et al., 1997), and gallocatechin is considered more antimicrobial than catechin (Scalbert, 1991). Moreover, it has been shown that prodelphinidin-rich PA polymers oxidize more rapidly at high pH than procyanidin-rich ones (Barbehenn et al., 2006a) and that prodelphinidin-rich polymers are able to better inhibit the oxidation of hydrolyzable tannins in these alkaline conditions than are procyanidin-rich polymers (Barbehenn et al., 2006b). In leaf litter in soils, prodelphinidin-rich PAs were more inhibitory to net nitrogen mineralization rates than those dominated by procyanidins (Nierop et al., 2006). The biological impact of B-ring hydroxylation could be tested directly using genetic modification in transgenic poplars; the genes we have identified as part of our study will be useful tools in this endeavor.
In summary, our characterization of MYB134 and MYB115 regulators of PAs in poplar emphasizes the complexity of the PA gene regulatory network and provides a novel suite of candidate genes for further study, including additional activator and repressor MYBs. Functional analysis of these MYBs, coupled to chemical analysis of PAs and other phenolics in transgenic plants, is a powerful approach to understanding gene function and has led to the discovery of new interactions within phenolic metabolism of poplar.
MATERIALS AND METHODS
Plant Growth Conditions and Treatments
Populus tremula × Populus tremuloides (clone INRA 353-38) plantlets were maintained and propagated in vitro on Murashige and Skoog (MS) medium with 0.5 µm indole-3-butyric acid (IBA). When plantlets had good root growth, they were transplanted to soil, acclimated in a mist chamber for 3 weeks, and planted in 1-gallon pots with a peat-based potting mix as described (Major and Constabel, 2006). Plants were maintained in the greenhouse with supplemental light (16-h days) between 18°C to 28°C. Plants were typically 3 months old with 20 to 25 leaves when used for experiments. For wounding, margins of leaves of LPI 10 to 12 (Larson and Isebrands, 1971) were crushed once with pliers and rewounded after 1 h. LPI 1 was defined as the first fully opened leaf with a lamina length of at least 1.5 cm. Wounded leaves were collected 24 h after initial wounding. Necrotic tissue and midveins were removed, and tissue was frozen in liquid nitrogen and stored at −80°C prior to analysis. For all analyses, leaves from LPI 10 to 12 were pooled, and at least three clones of each genotype or independent transgenic line were used.
Phylogenetic Analysis
A multiple alignment using coding sequences of plant MYB proteins related to phenylpropanoid synthesis was generated with ClustalW (2.0.12; Thompson et al., 1994). Nucleotide residues that were not highly conserved were trimmed manually in BioEdit Sequence Alignment Editor (version 7.2.5; Hall, 1999). The phylogenetic tree was generated by maximum likelihood using the fastDNAml package (version 1.2.2; Olsen et al., 1994). Bootstrap values were calculated from 1,000 phylogenetic constructions using a maximum likelihood analysis, and bootstrap values greater than 500 were mapped back onto the original tree. The graphical representation of the phylogenetic tree was generated with FigTree (version 1.4). Accession numbers were as follows: VvMYBA2, DQ886420.1; PtMYB120, XM_006383258; PtremMYB117, KP723394.1; PhAN2, AF146702.1; AtMYB114, AY008379.1; AtMYB113, AY519566.1; AtPAP2, AF062915.2; AtPAP1, AY519563; MtMYB5, XM_003601561.2; VvMYB5a, XM_002281607; AtMYB5, AY519587; AtMYB7, AY519573; AtMYB4, AF062860; AtMYB6, AY519604; AtMYB3, AY072543; PtMYB182, XM_002305836; PtMYB165, XM_002315854, PtMYB194, XM_002311459.2; AtMYB111, AF371977; AtMYB11, NM_116126; AtMYB12, AF062864; PtMYB115, XM_002302608.2; PtMYB201, XM_002320840.2; PtMYB153, XM_002303641.2; PtMYB123, XM_002304534.1; DkMYB4, AB503701.1; PpPA1, XM_008234893.1; VvMYBPA1, AM259485.1; AtTT2, AF371981.2; FaMYB11, JQ989282.1; MdMYB11, DQ074463.1; VvPA2, EU919682.1; DkMYB2, AB503699.1; ZmC1, NM_001112540.1; PtremMYB134, FJ573151.1; PtMYB087, XM_002324158.2; PtMYB086, XM_006371832; MdMYB9, DQ267900.1; FaMYB9, JQ989281.1; MtPAR, HQ337434.1; LjTT2a, AB300033.2; LjTT2c, AB300035.1; LjTT2b, AB300034.2; TaMYB14, JN049641.1; MtMYB14, XM_013602969; AtMYB23, AY519631; and AtGL1, AF495524.
Generation of Transgenic Plants Overexpressing PtMYB115
The complete sequence of MYB115 was PCR amplified from P. trichocarpa (Nisqually 1); Supplemental Table S5 and cloned into pBI526 prior to subcloning into the binary transformation vector pRD400 (Datla et al., 1993; Wang and Constabel, 2004). The pRD400:MYB115 plasmid was moved into Agrobacterium tumefaciens strain C58 (pMP90) by electroporation. P. tremula × P. alba (clone INRA 717-1B4) and P. tremula × P. tremuloides (clone INRA 353-38) were transformed using the method of Han et al. (2000) with minor modifications. A. tumefaciens carrying the appropriate plasmid was grown overnight with shaking at 225 rpm at 28°C. Cells were pelleted and resuspended in induction medium (MS medium with vitamins and 1.28 mm MES, 10 mm Gal, and 50 µm acetosyringone) to an OD600 of 0.5 and then incubated with shaking to an OD600 of 0.6. Explants were excised from 2-month-old in vitro plantlets, wounded with a sterile scalpel on the leaf surface, and incubated in the A. tumefaciens suspension for 1 h. The explants were then blotted to remove excess bacteria, transferred to solid callus induction medium 1 (CIM1) plates (MS medium with 5 µm 2-isopentenyl adenine and 10 µm α-naphthalene acetic acid), and incubated in the dark. After 2 d, explants were transferred to callus induction medium 2 plates (CIM1 with 250 mg L−1 cefotaxime, 500 mg L−1 carbenicillin, and 50 mg L−1 kanamycin) and incubated for 3 weeks in darkness. When calli appeared, explants were transferred to shoot induction medium (CIM1 with 250 mg L−1 cefotaxime, 500 mg L−1 carbenicillin, 50 mg L−1 kanamycin, plus 0.2 µm thidiazuron) and placed in growth chambers under low light (approximately 50 µE m−2 s−1). When shoots reached 0.5 to 1 cm in height, they were excised and transferred to root induction medium (one-half-strength MS medium with 1.25 µm IBA). Plants were screened for a high-tannin phenotype using a p-dimethylaminocinnamaldehyde stain (1% [w/v] in ethanol:6 n HCl, 1:1 [v/v]; Feucht and Treutter, 1990; Mellway et al., 2009). Transgenics were confirmed by PCR using primers for the 35S promoter and NOS terminator. Positive transformants were micropropagated on solid MS medium with 0.5 µm IBA.
Phytochemical Analysis
Extracts for phenolic quantitation were prepared with 25 mg of finely ground, freeze-dried tissue and extracted three times sequentially in a total volume of 4.5 mL of 100% HPLC-grade methanol by sonication. Extracts were centrifuged for 10 min at 13,000 rpm, and the supernatant was dried in an SC110A SpeedVac Plus concentrator. Dried extracts were resuspended in 300 µL of 100% methanol, and chlorophyll was removed using Strata-X 33-µm solid-phase extraction columns (Phenomenex). The column was conditioned with 3 mL of 100% methanol and then 3 mL of deionized water before applying sample and eluting phenolics with 9 mL of 100% methanol. Extracts were again dried in glass tubes in an SC110A SpeedVac Plus concentrator, weighed, and resuspended in 100% methanol to 10 mg mL−1. Each sample (20 µL) was analyzed using an HPLC system (Beckman Coulter System Gold 126 solvent module with a System Gold 168 diode array detector) equipped with a Phenomenex Kinetex C18 column (150 × 4.6, 2.6 µm; 100 Å). Separation was performed with an elution gradient using solvent A (deionized water with 0.4% [v/v] formic acid) to solvent B (acetonitrile with 0.4% [v/v] formic acid) over 55 min at a flow rate of 1 mL min−1. The gradient profile was 0% to 5% B (0–5 min), 5% to 14% B (5–11 min), 14% to 38% B (11–40 min), 38% to 100% B (40–47 min), and 5% B (47–49 min). Data analysis was performed with 35 Karat Software version 5.0 (Beckman Coulter). The baseline was manually added for the integration of peak area. Compounds of interest were quantified by UV detection at 280 nm using representative standards: phenolic glycosides were quantified as salicin equivalents, catechin and dihydromyricetin were quantified relative to the commercial standards (Sigma). Total PAs were quantified using the butanol-HCl method as described by Porter et al. (1986) using purified poplar tannins as a standard. Extracts (500 µL) were added to 2 mL of butanol-HCl (95:5, v/v) with 66.7 µL of iron reagent [2% (w/v) NH4Fe(SO4)2 in 2 n HCl] and heated at 95°C for 40 min. Absorbance was read at 550 nm and corrected for absorbance of corresponding unheated control samples. All statistical tests for metabolites were done in R (www.r-project.org). For ANOVA, the AOV function from the stats version 2.15.0 package was used.
Unknown differential peaks were further characterized by LC-MS. A Bruker Esquire 6000 ion-trap mass spectrometer (Bruker Daltonics) was operated in alternating ionization mode in the range m/z 60 to 1,000 (capillary exit voltage, +121/−121 electron volts; capillary voltage, +4,000/−4,000V; nebulizer pressure, 35 p.s.i.; drying gas, 11 L min−1; and gas temperature, 330°C) coupled to an Agilent 1100 series HPLC device (Agilent Technologies). Elution was accomplished using a Kinetics C18 column (100 × 4.6 mm, 2.6 µm; Phenomenex). Mobile phases were 0.2% (v/v) formic acid (A) and acetonitrile (B), starting with 5% B for 5 min, followed by a gradient: 5% to 14% B (5–11 min), 14% to 38% B (11–40 min), 100% B (40–43 min), and 5% B (43–47 min). Additional fragmentation for selected ions (MS2 and MS3 spectra in positive and negative modes) were obtained with the same instrument in AutoMS mode for the unknown compounds (m/z 423 and 319 in negative mode) and also the authentic standard for dihydromyricetin (Sigma).
For group-specific UPLC-MS/MS analysis of procyanidin- and prodelphinidin-containing PAs and three flavonol subgroups (kaempferol, quercetin, and myricetin glycosides), 20 mg of finely ground leaf powder was extracted with 3 × 1.4 mL of acetone:water (80:20, v/v) on a rotary shaker for 3 × 3 h (280 rpm), followed by centrifugation for 10 min. The supernatant was transferred to a new microcentrifuge tube, and acetone was removed in an Eppendorf concentrator (5301). Aqueous samples were frozen at –20°C and lyophilized. The freeze-dried phenolic extract was resuspended in 1 mL of Milli-Q purified water, vortexed for 5 min, and filtered with a 0.2-µm polytetrafluoroethylene filter into UPLC vials. The analyses were conducted according to Engström et al. (2014) on an Acquity UPLC system (Waters) interfaced to a Xevo TQ triple-quadrupole mass spectrometer with electrospray ionization (ESI; Waters). The UPLC system was equipped with an autosampler, a binary solvent manager, a 100-mm × 2.1-mm i.d., 1.7-μm Acquity UPLC BEH Phenyl column (Waters), and a diode array detector. The flow rate was set to 0.5 mL min−1, and the mobile phase consisted of two solvents, acetonitrile (A) and 0.1% aqueous formic acid (B), with the following gradient: 0.1% A in B (0−0.5 min, isocratic), 0.1% to 30% A in B (0.5−5 min, linear gradient), 30% to 35% A in B (5−6 min, linear gradient), column wash, and stabilization (6−9.5 min). Data collection of both UV and MS medium occurred continuously from 0 to 6 min. Negative ESI mode was used, with ESI conditions as follows: capillary voltage, 2.4 kV; desolvation temperature, 650°C; source temperature, 150°C; desolvation and cone gas (N2), 1,000 and 100 L h−1, respectively; and argon as collision gas. Five replicates of a stock solution of catechin (1 µg mL−1) were analyzed every 2 h by a catechin-specific MRM method to account for possible changes in the quantitative performance of the system. In addition, a mixture of five flavonoid glycosides was analyzed before every sample batch to ensure the stability of the retention times and m/z positions between different batches. Calibration curves (1,000–1 µg mL−1) for procyanidin and prodelphinidin concentrations were produced from Sephadex LH-20 purified procyanidin-rich and prodelphinidin-rich fractions. Under our conditions, the limit of detection was 0.0085 mg g−1 dry weight for prodelphinidin and 0.0008 mg g−1 dry weight for procyanidin (Engström et al., 2014). Calibration curves (8–0.1 µg mL−1) for kaempferol, quercetin, and myricetin glycosides were produced from kaempferol-3-O-glucoside, quercetin-3-O-galactoside, and myricetin-3-O-rhamnoside, respectively.
RNA Extraction and RT-qPCR
RNA was extracted from approximately 50 mg of frozen ground leaf tissue using the method of Muoki et al. (2012). For quantitative PCR analysis, total RNA was treated with Amplification Grade DNase I (Invitrogen) to remove genomic DNA, and cDNA was generated using SuperScript II (Invitrogen). Expression was normalized against the poplar elongation factor 1-β (accession no. XM_002299894). Quantitative PCR was carried out with the QuantiTect SYBR Green RT-PCR kit (Qiagen). Each reaction contained 5 ng (2 µL) of a 1:20 diluted cDNA template, 1 µL of 10 µm forward and reverse primers (Table II), 7.5 µL of 2× QuantiTect master mix, and 4.5 µL of water in a total volume of 15 µL. No-reverse transcriptase controls were included for each sample, and amplification was performed on the Mx3005p QPCR System (Stratagene). The PCR conditions were as follows: 10 min at 95°C followed by 40 cycles of 95°C for 30 s, 56°C to 60°C for 30 s, and 72°C for 30 s, followed by one cycle of 95°C for 1 min, 56°C to 60°C for 30 s, and 95°C for 30 s. For each sample run, a melt-curve analysis was performed using the Mx3005p default parameters (60 s at 95°C, 30 s at 55°C–95°C in 1°C increments, and 30 s at 95°C), which yielded one peak after normalization with the carboxy-X-rhodamine (ROX) reference dye ROX signal for each set of primers. Annealing temperatures were optimized for highest primer efficiency. Primer efficiency was estimated by calculating the slope of a dilution series of template concentrations. Efficiency was calculated using the slope in the following equation: primer efficiency (%) = ((10−(1/slope)) − 1)100.
Microarray Analysis
RNA extraction was performed as described above with additional RNA cleanup using the NucleoSpin RNA II cleanup kit (Clontech). Affymetrix GeneChip Poplar Genome Array microarray hybridizations were conducted at the Genome Quebec Innovation Centre at McGill University. Data were normalized with FlexArray (http://www.gqinnovationcenter.com/downloads/index.aspx?l=e) using a robust multiarray average algorithm. To identify differentially expressed genes, the Empirical Bayes (Wright and Simon, 2003) algorithm was performed in FlexArray. Statistical analysis was performed with the R program (http://www.r-project.org/) using BioConductor (Gentleman et al., 2004). Genes with a fold change greater than 2 or less than 0.5 (MYB overexpressors versus controls) at P ≤ 0.05 were defined as differentially expressed. Annotations were obtained from annotation files provided by Affymetrix. Gene model identifiers were looked up at POParray (http://aspendb.uga.edu) and further annotations were obtained from Blast2Go analysis (Conesa et al., 2005) and MapMan (http://mapman.gabipd.org/). Annotations were further verified by BLASTN searches in the National Center for Biotechnology Information Transcript Reference Sequence Database and by keyword searches in Phytozome version 9.1 (http://phytozome.jgi.doe.gov; P. trichocarpa Joint Genome Institute assembly, release version 3.0, annotation version 3.0). For genes with duplicate probe sets, the one with the highest fold change was used. All materials and procedures complied with Minimum Information About a Microarray Experiment standards set for array data (Brazma et al., 2001).
Transient Plant Transformation by Particle Bombardment for Promoter Activation Assays
For dual-luciferase reporter assays, pMDC32 overexpression vectors (Curtis and Grossniklaus, 2003; Hellens et al., 2005) and the pGREEN800LUC reporter vector (Hellens et al., 2005) were used. Promoter fragments and cDNAs were cloned into pGEM T-Easy (Promega) and then subcloned into the NotI sites of pGREEN800LUC (promoters) or pMDC32 (transcription factors). Promoter regions were between 1 and 2 kb upstream of the gene of interest (for primer list, see Supplemental Table S5). For overexpression, sequences were cloned into the NotI sites of a pENTR and then recombined into pMDC32 vectors using the Gateway system (LR Clonase; Invitrogen).
P. trichocarpa × P. deltoides (H11-11) cells were maintained in liquid MS medium on a rotary shaker and subcultured every 8 to 10 d. Cells were bombarded on day 7 of the culture cycle. Particle bombardment was carried out with gold or tungsten beads coated with 250 ng of each plasmid. Prior to bombardment, poplar suspension cultures were allowed to settle for 15 min and the liquid medium was removed. Suspension cell aliquots were taken from four independently grown cultures and pipetted onto four individual filter papers for each bombardment. The filter paper with the cells was placed on solid MS medium (with 0.5 m mannitol), and the cells were incubated for 1 h prior to bombardment. All four replicates were bombarded simultaneously 13 cm from the rupture disc. Each vector (250 ng each of the activator, cofactor, and reporter vectors) was added to 25 µL of 0.6-µm gold or 0.7-µm tungsten particles (Bio-Rad) under constant vortexing, followed by 25 µL of 2.5 m CaCl2 and 10 μL of 0.1 m spermidine. Samples were further vortexed for an additional 10 min at 4°C. The beads were washed with 70% and 100% ethanol and resuspended in 20 µL of 100% ethanol. The beads were pipetted onto presterilized flying disks (Bio-Rad) and allowed to air dry. Rupture disks (900 p.s.i.) were used for the bombardments in the Bio-Rad model PDS-1000/He Biolistic Particle Delivery System. Following bombardment, the filters with cells were maintained on MS medium (0.5 m mannitol) plates in darkness for 48 h prior to measuring luciferase activity using the dual-luciferase reporter assay kit (Promega).
Yeast Two-Hybrid Assays
The Clontech Matchmaker Gold Yeast Two-Hybrid System was used to test for MYB-bHLH interactions. The N-terminal DNA binding domains of MYB134 and MYB115 were fused to the Gal4 binding domain in the pGBKT7 vector, and bHLH131 was fused to the Gal4 activation domain in the pGADT7 vector. Combinations of Gal4 binding and activation domain vectors were transformed into the Y2HGold yeast strain following the manufacturer's instructions (www.clontech.com/xxclt_ibcGetAttachment.jsp?cItemId=17597). Yeast cells containing both vectors were selected and tested on synthetically defined (SD) media lacking Leu, Trp, adenine or His (SD/-Leu/-Trp/-Ade/-His), as well as SD media without Leu or Trp plus 200ng/ml of Aureobasidine A (AbA) (SD/-Leu/-Trp/200ng/ml AbA). Serial dilutions of transformed yeast cells were carried out with 0.9% saline solution and tested on SD/-Leu/-Trp/200ng/ml AbA media.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Protein sequence alignment of MYB115 with other PA1 clade MYB activators.
Supplemental Figure S2. Analysis of the in silico expression of MYB115, MYB134, and other major PA pathway genes.
Supplemental Figure S3. RT-qPCR expression profiles of MYB115 and MYB134 in different poplar tissues.
Supplemental Figure S4. Expression of the MYB115 transgene in MYB115-overexpressing poplar lines.
Supplemental Figure S5. Validation of microarray using RT-qPCR.
Supplemental Figure S6. Overlay of sample HPLC profiles comparing MYB115-overexpressing and control poplar leaf extracts.
Supplemental Figure S7. Sample chromatograms showing fingerprints of procyanidin and prodelphinidin subunits in UPLC-MS/MS analysis.
Supplemental Table S1. List of transcripts significantly enriched in MYB134 overexpressors.
Supplemental Table S2. List of transcripts significantly enriched in MYB115 overexpressors.
Supplemental Table S3. Analysis of PAs in MYB-overexpressing plants.
Supplemental Table S4. Concentrations of kaempferol, quercetin, and myricetin glycosides in MYB134- and MYB115-overexpressing poplars.
Supplemental Table S5. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Brad Binges and the University of Victoria Centre for Forest Biology for help with plant care and growth facilities, Oliver Corea for help with gene annotations, and Marie-Josée Morency (Laurentian Forestry Centre) for help in poplar gene expression analyses.
Glossary
- PA
proanthocyanidin
- RT-qPCR
reverse transcription-quantitative PCR
- LC-MS
liquid chromatography-mass spectrometry
- MS/MS
tandem mass spectrometry
- UPLC
ultra-performance liquid chromatography
- MRM
multiple reaction monitoring
- IBA
indole-3-butyric acid
- MS
Murashige and Skoog
- LPI
leaf plastochron index
- CIM1
callus induction medium 1
- ESI
electrospray ionization
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada, the Max Planck Society, the Academy of Finland, and the Canadian Genomics R&D Initiative.
Articles can be viewed without a subscription.
References
- Akagi T, Ikegami A, Tsujimoto T, Kobayashi S, Sato A, Kono A, Yonemori K (2009) DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol 151: 2028–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Ikegami A, Yonemori K (2010) DkMyb2 wound-induced transcription factor of persimmon (Diospyros kaki Thunb.), contributes to proanthocyanidin regulation. Planta 232: 1045–1059 [DOI] [PubMed] [Google Scholar]
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26: 962–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres MP, Clausen TP, MacLean SF, Redman AM, Reichardt PB (1997) Diversity of structure and antiherbivore activity in condensed tannins. Ecology 78: 1696–1712 [Google Scholar]
- Barbehenn RV, Constabel CP (2011) Tannins in plant-herbivore interactions. Phytochemistry 72: 1551–1565 [DOI] [PubMed] [Google Scholar]
- Barbehenn RV, Jones CP, Hagerman AE, Karonen M, Salminen JP (2006a) Ellagitannins have greater oxidative activities than condensed tannins and galloyl glucoses at high pH: potential impact on caterpillars. J Chem Ecol 32: 2253–2267 [DOI] [PubMed] [Google Scholar]
- Barbehenn RV, Jones CP, Karonen M, Salminen JP (2006b) Tannin composition affects the oxidative activities of tree leaves. J Chem Ecol 32: 2235–2251 [DOI] [PubMed] [Google Scholar]
- Baudry A, Caboche M, Lepiniec L (2006) TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J 46: 768–779 [DOI] [PubMed] [Google Scholar]
- Boeckler GA, Gershenzon J, Unsicker SB (2011) Phenolic glycosides of the Salicaceae and their role as anti-herbivore defenses. Phytochemistry 72: 1497–1509 [DOI] [PubMed] [Google Scholar]
- Boeckler GA, Towns M, Unsicker SB, Mellway RD, Yip L, Hilke I, Gershenzon J, Constabel CP (2014) Transgenic upregulation of the condensed tannin pathway in poplar leads to a dramatic shift in leaf palatability for two tree-feeding Lepidoptera. J Chem Ecol 40: 150–158 [DOI] [PubMed] [Google Scholar]
- Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. (2001) Minimum information about a microarray experiment (MIAME): toward standards for microarray data. Nat Genet 29: 365–371 [DOI] [PubMed] [Google Scholar]
- Broun P. (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol 8: 272–279 [DOI] [PubMed] [Google Scholar]
- Cavallini E, Matus JT, Finezzo L, Zenoni S, Loyola R, Guzzo F, Schlechter R, Ageorges A, Arce-Johnson P, Tornielli GB (2015) The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol 167: 1448–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedgy RJ, Köllner TG, Constabel CP (2015) Functional characterization of two acyltransferases from Populus trichocarpa capable of synthesizing benzyl benzoate and salicyl benzoate, potential intermediates in salicinoid phenolic glycoside biosynthesis. Phytochemistry 113: 149–159 [DOI] [PubMed] [Google Scholar]
- Close DC, McArthur C (2002) Rethinking the role of many plant phenolics: protection from photodamage not herbivores? Oikos 99: 166–172 [Google Scholar]
- Coleman HD, Park JY, Nair R, Chapple C, Mansfield SD (2008) RNAi-mediated suppression of p-coumaroyl-CoA 3′-hydroxylase in hybrid poplar impacts lignin deposition and soluble secondary metabolism. Proc Natl Acad Sci USA 105: 4501–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datla RSS, Bekkaoui F, Hammerlindl JK, Pilate G, Dunstan DI, Crosby WL (1993) Improved high-level constitutive foreign gene-expression in plants using an AMV RNA4 untranslated leader sequence. Plant Sci 94: 139–149 [Google Scholar]
- de Vetten N, ter Horst J, van Schaik HP, de Boer A, Mol J, Koes R (1999) A cytochrome b5 is required for full activity of flavonoid 3′,5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc Natl Acad Sci USA 96: 778–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Xie DY, Sharma SB (2005) Proanthocyanidins: a final frontier in flavonoid research? New Phytol 165: 9–28 [DOI] [PubMed] [Google Scholar]
- Engström MT, Pälijärvi M, Fryganas C, Grabber JH, Mueller-Harvey I, Salminen JP (2014) Rapid qualitative and quantitative analyses of proanthocyanidin oligomers and polymers by UPLC-MS/MS. J Agric Food Chem 62: 3390–3399 [DOI] [PubMed] [Google Scholar]
- Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, et al. (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21: 168–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feucht W, Treutter D (1990) Flavan-3-ols in trichomes, pistils and phelloderm of some tree species. Ann Bot (Lond) 65: 225–230 [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesell A, Yoshida K, Tran LT, Constabel CP (2014) Characterization of an apple TT2-type R2R3 MYB transcription factor functionally similar to the poplar proanthocyanidin regulator PtMYB134. Planta 240: 497–511 [DOI] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Hamberger B, Ellis M, Friedmann M, Souza CDA, Barbazuk B, Douglas CJ (2007) Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: the Populus lignin toolbox and conservation and diversification of gene families. Can J Bot 85: 1182–1201 [Google Scholar]
- Han KH, Meilan R, Ma C, Strauss SH (2000) An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep 19: 315–320 [DOI] [PubMed] [Google Scholar]
- Hancock KR, Collette V, Fraser K, Greig M, Xue H, Richardson K, Jones C, Rasmussen S (2012) Expression of the R2R3-MYB transcription factor TaMYB14 from Trifolium arvense activates proanthocyanidin biosynthesis in the legumes Trifolium repens and Medicago sativa. Plant Physiol 159: 1204–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton MK, Lindroth RL, Nordheim EV (2003) Foliar quality influences tree-herbivore-parasitoid interactions: effects of elevated CO2, O3, and plant genotype. Oecologia 137: 233–244 [DOI] [PubMed] [Google Scholar]
- Huang YF, Vialet S, Guiraud JL, Torregrosa L, Bertrand Y, Cheynier V, This P, Terrier N (2014) A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol 201: 795–809 [DOI] [PubMed] [Google Scholar]
- Hwang SY, Lindroth RL (1997) Clonal variation in foliar chemistry of aspen: effects on gypsy moths and forest tent caterpillars. Oecologia 111: 99–108 [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Larson PR, Isebrands JG (1971) The plastochron index as applied to developmental studies of cottonwood. Can J Res 1: 1–11 [Google Scholar]
- Liu J, Osbourn A, Ma P (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant 8: 689–708 [DOI] [PubMed] [Google Scholar]
- Major IT, Constabel CP (2006) Molecular analysis of poplar defense against herbivory: comparison of wound- and insect elicitor-induced gene expression. New Phytol 172: 617–635 [DOI] [PubMed] [Google Scholar]
- Malisch CS, Lüscher A, Baert N, Engström MT, Studer B, Fryganas C, Suter D, Mueller-Harvey I, Salminen JP (2015) Large variability of proanthocyanidin content and composition in sainfoin (Onobrychis viciifolia). J Agric Food Chem 63: 10234–10242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Alon U (2003) Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA 100: 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinova K, Pourcel L, Weder B, Schwarz M, Barron D, Routaboul JM, Debeaujon I, Klein M (2007) The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19: 2023–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellway RD, Tran LT, Prouse MB, Campbell MM, Constabel CP (2009) The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol 150: 924–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min BR, Barry TN, Attwood GT, McNabb WC (2003) The effect of condensed tannins on the nutrition and health of ruminants fed fresh temperate forages: a review. Anim Feed Sci Technol 106: 3–19 [Google Scholar]
- Miranda M, Ralph SG, Mellway R, White R, Heath MC, Bohlmann J, Constabel CP (2007) The transcriptional response of hybrid poplar (Populus trichocarpa x P. deltoides) to infection by Melampsora medusae leaf rust involves induction of flavonoid pathway genes leading to the accumulation of proanthocyanidins. Mol Plant Microbe Interact 20: 816–831 [DOI] [PubMed] [Google Scholar]
- Muoki RC, Paul A, Kumari A, Singh K, Kumar S (2012) An improved protocol for the isolation of RNA from roots of tea (Camellia sinensis (L.) O. Kuntze). Mol Biotechnol 52: 82–88 [DOI] [PubMed] [Google Scholar]
- Nierop KGJ, Preston CM, Verstraten JM (2006) Linking the B ring hydroxylation pattern of condensed tannins to C, N and P mineralization. A case study using four tannins. Soil Biol & Bioch 38: 2794–2802 [Google Scholar]
- Novobilský A, Stringano E, Hayot Carbonero C, Smith LM, Enemark HL, Mueller-Harvey I, Thamsborg SM (2013) In vitro effects of extracts and purified tannins of sainfoin (Onobrychis viciifolia) against two cattle nematodes. Vet Parasitol 196: 532–537 [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Matsuda H, Hagstrom R, Overbeek R (1994) fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci 10: 41–48 [DOI] [PubMed] [Google Scholar]
- Osawa H, Endo I, Hara Y, Matsushima Y, Tange T (2011) Transient proliferation of proanthocyanidin-accumulating cells on the epidermal apex contributes to highly aluminum-resistant root elongation in camphor tree. Plant Physiol 155: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osier TL, Lindroth RL (2006) Genotype and environment determine allocation to and costs of resistance in quaking aspen. Oecologia 148: 293–303 [DOI] [PubMed] [Google Scholar]
- Pearl IA, Darling SF (1969) Investigation of hot water extractives of Populus balsamifera bark. Phytochemistry 8: 2393–2396 [Google Scholar]
- Pearl IA, Darling SF (1971) Studies on leaves of family Salicaceae. 16. Phenolic extractives of leaves of Populus balsamifera and of P. trichocarpa. Phytochemistry 10: 2844–2847 [Google Scholar]
- Peters DJ, Constabel CP (2002) Molecular analysis of herbivore-induced condensed tannin synthesis: cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J 32: 701–712 [DOI] [PubMed] [Google Scholar]
- Porter LJ, Hrstich LN, Chan BG (1986) The conversion of procyanidins and prodelphinidins to cyanidin and delphidin. Phytochemistry 25: 223–230 [Google Scholar]
- Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10: 63–70 [DOI] [PubMed] [Google Scholar]
- Ravaglia D, Espley RV, Henry-Kirk RA, Andreotti C, Ziosi V, Hellens RP, Costa G, Allan AC (2013) Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol 13: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault H, Bassard JE, Hamberger B, Werck-Reichhart D (2014) Cytochrome P450-mediated metabolic engineering: current progress and future challenges. Curr Opin Plant Biol 19: 27–34 [DOI] [PubMed] [Google Scholar]
- Salminen JP, Karonen M (2011) Chemical ecology of tannins and other phenolics: we need a change in approach. Funct Ecol 25: 325–338 [Google Scholar]
- Salminen JP, Karonen M, Sinkkonen J (2011) Chemical ecology of tannins: recent developments in tannin chemistry reveal new structures and structure-activity patterns. Chemistry 17: 2806–2816 [DOI] [PubMed] [Google Scholar]
- Scalbert A. (1991) Antimicrobial properties of tannins. Phytochemistry 30: 3875–3883 [Google Scholar]
- Schaart JG, Dubos C, Romero De La Fuente I, van Houwelingen AMML, de Vos RCH, Jonker HH, Xu W, Routaboul JM, Lepiniec L, Bovy AG (2013) Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol 197: 454–467 [DOI] [PubMed] [Google Scholar]
- Schweitzer JA, Madritch MD, Bailey JK, LeRoy CJ, Fischer DG, Rehill BJ, Lindroth RL, Hagerman AE, Wooley SC, Hart SC, et al. (2008) From genes to ecosystems: the genetic basis of condensed tannins and their role in nutrient regulation in a Populus model system. Ecosystems (N Y) 11: 1005–1020 [Google Scholar]
- Scioneaux AN, Schmidt MA, Moore MA, Lindroth RL, Wooley SC, Hagerman AE (2011) Qualitative variation in proanthocyanidin composition of Populus species and hybrids: genetics is the key. J Chem Ecol 37: 57–70 [DOI] [PubMed] [Google Scholar]
- Stringano E, Hayot Carbonero C, Smith LMJ, Brown RH, Mueller-Harvey I (2012) Proanthocyanidin diversity in the EU ‘HealthyHay’ sainfoin (Onobrychis viciifolia) germplasm collection. Phytochemistry 77: 197–208 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Brugliera F (2013) Flower colour and cytochromes P450. Philos Trans R Soc Lond B Biol Sci 368: 20120432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier N, Torregrosa L, Ageorges A, Vialet S, Verriès C, Cheynier V, Romieu C (2009) Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol 149: 1028–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan Y (2006) Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol 172: 47–62 [DOI] [PubMed] [Google Scholar]
- Wang J, Constabel CP (2004) Polyphenol oxidase overexpression in transgenic Populus enhances resistance to herbivory by forest tent caterpillar (Malacosoma disstria). Planta 220: 87–96 [DOI] [PubMed] [Google Scholar]
- Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM (2009) Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol 149: 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GW, Simon RM (2003) A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 19: 2448–2455 [DOI] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396–399 [DOI] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20: 176–185 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Iwasaka R, Kaneko T, Sato S, Tabata S, Sakuta M (2008) Functional differentiation of Lotus japonicus TT2s, R2R3-MYB transcription factors comprising a multigene family. Plant Cell Physiol 49: 157–169 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Ma D, Constabel CP (2015) The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol 167: 693–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Pang Y, Dixon RA (2010) The mysteries of proanthocyanidin transport and polymerization. Plant Physiol 153: 437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zifkin M, Jin A, Ozga JA, Zaharia LI, Schernthaner JP, Gesell A, Abrams SR, Kennedy JA, Constabel CP (2012) Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol 158: 200–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40: 22–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.