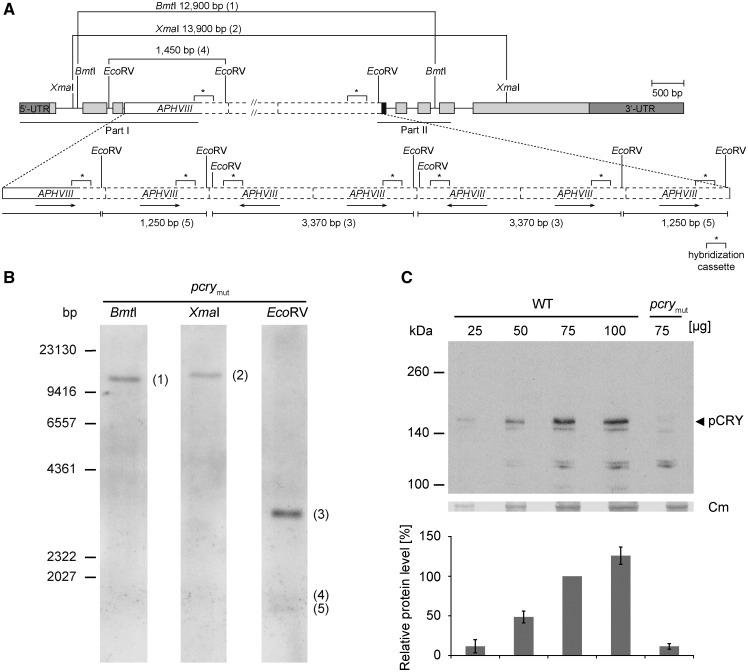
Figure 1.
Characterization of a Chlamydomonas pcry mutant. A, The gene model of pcry in the insertional mutant CRMS102 is shown. This genetic scheme depicts one of the possibilities that are in agreement with the results from DNA sequencing (Supplemental Fig. S1) and Southern-blot analysis (in B). Boxes in light gray show exons, and black lines indicate introns. Boxes in dark gray depict the 5′ and 3′ untranslated regions. The black box indicates an insertion of 60 bp of unknown origin. The APHVIII cassette (white box) appears to be inserted seven times in intron 3 with different orientations (black arrows). Solid lines of the white box indicate sequenced parts along with small insertions or deletions, and dashed lines are nonsequenced regions. The labeled sequence-specific probe (299 bp) in the cassette used for Southern blotting is depicted as asterisks with square brackets. Sequenced regions in pCRY and the APHVIII cassette are indicated by black lines (middle, according to part I and part II in Supplemental Fig. S1C). Relevant restriction sites are shown. B, Southern-blot analysis of CRMS102 to determine the number of insertions of the APHVIII cassette. Genomic DNA (30 µg per lane) from the pcry mutant strain was digested with XmaI, BmtI, or EcoRV, as indicated. DNA fragments were separated on a 0.8% agarose gel and blotted onto a nylon membrane. For hybridization, a digoxigenin-labeled DNA fragment of the APHVIII cassette was used (see “Materials and Methods”). It was shown before that this probe does not give a signal with wild-type genomic DNA (Beel et al., 2012). C, Quantification of pCRY levels in the pcry insertional mutant. Cells were grown under an LD12:12 cycle and harvested in the night (LD22). Different amounts of proteins from crude extracts (25, 50, 75, and 100 µg per lane) of SAG73.72 wild-type (WT) and pcrymut cells were separated by 7% SDS-PAGE and used for immunoblots with anti-pCRY antibodies. The position of pCRY is indicated by the arrowhead. As a loading control, the polyvinylidene difluoride (PVDF) membrane was stained with Coomassie Brilliant Blue R 250 after immunochemical detection. From this stain, selected unspecified protein bands are shown (Cm). The quantified pCRY protein levels of three biological replicates are shown in the diagram at bottom.