Abstract
Post-transcriptional processes orchestrate gene expression through dynamic protein-RNA interactions. These interactions occur at specific sites determined by RNA sequence, secondary structure, or nucleotide modifications. Methods have been developed either to quantify binding of whole transcripts or to identify the binding sites, but there is none proven to quantify binding at both the whole transcript and binding site levels. Here we describe digestion optimized RNA immunoprecipitation with deep sequencing (DO-RIP-seq) as a method that quantitates at the whole transcript target (RIP-Seq-Like or RSL) level and at the binding site level (BSL) using continuous metrics. DO-RIP-seq methodology was developed using the RBP HuR/ELAVL1 as a test case (Nicholson et al. 2016). DO-RIP-seq employs treatment of cell lysates with a nuclease under optimized conditions to yield partially digested RNA fragments bound by RNA binding proteins, followed by immunoprecipitations that capture the digested RNA-protein complexes and assess non-specific or background interactions. Analyses of sequenced cDNA libraries made from the bound RNA fragments yielded two types of enrichment scores; one for RSL binding events and the other for BSL events (Nicholson et al. 2016). These analyses plus the extensive read coverage of DO-RIP-seq allows seamless integration of binding site and whole transcript information. Therefore, DO-RIP-seq is useful for quantifying RBP binding events that are regulated during dynamic biological processes.
Keywords: DO-RIP-seq, RNA-binding proteins, RNA-binding sites, whole-transcript targets, quantitation
1. Introduction
RNA-protein interactions mark sites and regions of post-transcriptional regulation of gene expression. RNA-binding proteins (RBPs) and noncoding RNAs govern the functions of mRNAs by binding to specific RNA sequences, structures or chemical modifications. Knowledge of precise RNA binding sites can illuminate combinatorial interactions between RBPs and non-coding RNA, the determinants of ribonucleoprotein assembly, and functional outcomes at the whole transcript level. Therefore, it is crucial to develop methods that quantitatively identify the relative binding strength of mRNA targets of RBPs in the cell across the transcriptome.
The first transcriptome wide RIP (RNA-binding protein immunoprecipitation) method, RIP-Chip, was originally developed in our lab to identify RNAs targeted by a RBP [1] and has since been modified, refined and applied by numerous labs to a wide variety of problems (reviewed in [2]). Our RIP method calculates probabilities of RBP-RNA association by normalizing the abundance of RNA isolated by immunoprecipitation (IP) of a RBP to the abundance in the negative IP (or background). The probability scores that result can be continuous metrics of RBP-RNA association that quantify changes in the association that result in RNP remodeling during dynamic biological conditions [1, 3-6]. However, RIP does not yield the binding sites of the RBP along the target RNAs.
Global RIP-crosslinking procedures whereby RBPs and RNAs are crosslinked in cells were also invented in our lab [7] and applied by Darnell and coworkers to identify mRNA targets [8]. The identification of binding sites was achieved by using biochemical and computational adaptations to these crosslinking immunoprecipitation (CLIP) procedures [9-12]. However, the CLIP method as initially practiced did not integrate background-binding measurements into the identification of binding sites. This is because it was assumed that the stringent conditions used to separate crosslinked protein-RNA complexes would remove all background. It has since been shown that considerable crosslinking background remains in CLIP preparations, but the background can be used to improve “binding site calling” and reduces false positives when properly applied [13]. However, despite the incorporation of background binding measurements, CLIP procedures are unable to quantify binding events largely because of the low efficiency of UV crosslinking [14]. Nevertheless, RIP and CLIP can provide useful complementary information despite their differences [15-24], and therefore, we sought to develop a procedure that uses aspects of both protocols to quantify whole transcript targets as well as binding sites.
Here we outline the DO-RIP-seq protocol for transcriptome-wide quantification of RNA binding sites for RBPs. DO-RIP-seq melds features of both RIP and CLIP procedures to measure enrichment scores for binding to both whole transcripts and binding sites. This is achieved by treating protein-RNA complexes in cell lysates with nucleases; in this case, micrococcal nuclease under conditions optimized for partial digestion (Fig. 1). This step is followed by immunoprecipitations with antibodies against the RBP of interest, while using non-specific antibodies in parallel to measure background and account for overall transcript abundance. As described here, DO-RIP-seq was developed using the RBP ELAVL1/HuR as a test case, and the experiments yielded probabilistic measures (log of odds scores, LOD) for binding sites in HEK293 cells [25], and for XIST long non-coding RNA in mouse trophoblasts [26]. HuR binding site LOD scores correlated with binding strength and motif preference [25]. Also DO-RIP-seq analysis of whole transcript association distinguished functional groups of messages and enriched gene sets [25]. In addition, we have used DORIP-seq to successfully identify the binding sites of other RBPs, e.g. RBM38, CELF1, and TRA2B (unpublished works in progress). Therefore, DO-RIP-seq provides continuous metrics of RBP targeting at the resolution of the whole transcript and the binding site, and thus, allows one to connect cumulative binding site metrics to functional outcomes at the whole transcript level. Note that while our presentation of the protocol is exceedingly detailed, the procedure is relatively straightforward, rapid, and user friendly.
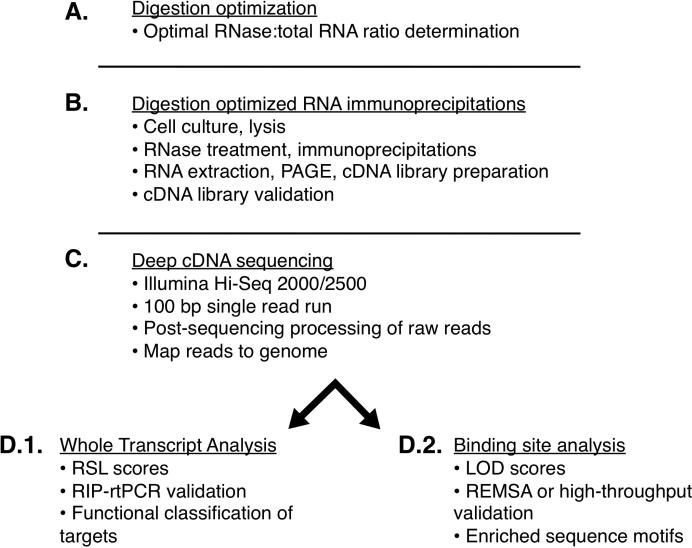
Figure 1. The DO-RIP-seq workflow.
A. Determine the optimal quantitative ratio of RNase to total RNA for obtaining protein-bound RNA fragments for mapping binding sites. B. Digestion optimized RNA immunoprecipitations of protein-bound and non-specifically bound (background) RNA fragments, and preparation of cDNA libraries from the extracted RNA. C. Sequencing of cDNA libraries on Illumina Hi-Seq 2000/2500 using 100 base pair (bp) single read runs. Processing of raw sequenced reads to remove oligonucleotide adapters and unique molecular identifiers, and then mapping to the appropriate species genome. D.1. Whole transcript analysis to calculate enrichment scores (RSL, RIP-seq-like) for expressed genes. RSL scores can be validated by RIP-rtPCR, and used as criteria for functional classification of targets. D.2. Binding site analysis to generate log of odds (LOD) scores which quantifies the probability of a site being bound by the protein relative to the site being in the background. Binding sites can be validated using REMSA or compared to in vitro high-throughput data. Also the binding sites can be used to discover enriched sequence motifs.
2. Materials & Methods
2.1. Buffers
2.1.1. Polysome lysis buffer
Prepare polysome lysis buffer with the following components in distilled, nuclease-free water and store it at 4 °C:
10 mM HEPES pH 7.0
100 mM KCl
5 mM MgCl2
5 mM CaCl2
0.5% (v/v) IGEPAL CA-630
Add the following components to the polysome lysis buffer when cells are ready for harvesting:
1 mM Dithiothreitol (DTT)
1X cOmplete™ protease inhibitor (Roche)
100 Units/ml RNaseOUT (Thermo Fisher Scientific)
If necessary greater lysis of certain cell types can be achieved by keeping the magnesium and calcium salts out of the lysis buffer. Magnesium is believed to have a stabilizing effect on membranes through electrostatic interactions with the negatively charged groups of the membranes [27]. We have found that leaving both magnesium and calcium out of the lysis buffers increases lysis efficiency for some cell types and this should be empirically determined in each case (not shown). However, these salts should be added to the lysates before treating the lysates with micrococcal nuclease. Micrococcal nuclease requires Ca2+ for activity [28], and Mg2+ is important for stabilizing RNA structures [29].
2.1.2. NT2 buffer
Prepare NT2 buffer in distilled, nuclease-free water using the following components and store it at 4 °C:
50 mM Tris-HCl pH 7.4
1 mM MgCl2
150 mM NaCl
0.05% (v/v) IGEPAL
2.2. Cell culture and lysate preparation
A single DO-RIP-seq experiment will require enough cell lysate for at least two immunoprecipitations (IPs); one IP with antibodies against the RBP of interest, and another using non-specific antibodies to measure background. The non-specific antibody we prefer is normal serum, for example normal mouse serum (Jackson ImmunoResearch Laboratories, cat. No. 015-000-001, see section 3.3). Normal serum from mouse is used as a negative control when the antibody used to immunoprecipitate the RBP is from mouse as well. Therefore, antibodies used to immunoprecipitate the RBP and to perform negative control IPs should be from matching species. The number of cells required for DO-RIP-seq experiments will depend on the abundance of the RBP in the lysate. We recommend starting with up to five 15-cm dishes of cells that are 80-90% confluent (approximately 12 × 106 cells per dish for HEK293 cell line) for each IP if possible. In our experience one 15-cm dish of HEK293 cells per IP is sufficient for DO-RIP-seq experiments done with antibodies against endogenous HuR/ELAVL1. While these amounts are ideal, smaller amounts have been used successfully in other cases.
Harvest cells by first removing the culture media from dish of cells, adding 2 ml of cold 1X PBS, and then scraping the cells from the surface of the dish using a Corning® cell lifter (product no. 3008) or similar tool. Next, transfer the scraped cells to a 15-ml centrifuge tube (or 50-ml centrifuge tubes if necessary), pooling like cells. Centrifuge for 5 min at 1200 rpm (speed may vary depending on cell type) at 4 °C. Discard the supernatant, and then wash the cell pellet with 10 ml of cold 1X PBS. Centrifuge the cells for 5 min at 1200 rpm and then discard the supernatant. It is important to remove as much residual PBS as possible because it interferes with the inactivation of micrococcal nuclease downstream. Resuspend the cell pellet in 1.5X cell pellet volumes of polysome lysis buffer (PLB) supplemented as described in sub-section 2.1.1 and transfer them to a clean 1.5 ml microcentrifuge tube. Repeatedly pipette the cells on ice with a P-1000 10-20 times, incubate the cell lysates on ice for 5-10 min, and then freeze the cell lysates at −80 °C. Prolonged freezing improves lysis, and the lysates can remain frozen for several months until ready for use. We recommend checking the lysis of your cell type upon thawing by observing 10 μl on glass slides, under a light microscope. Some cell types may require pumping the cells through a small gauge needle to obtain good lysis, but note that sonication should NOT be used.
2.3. Preparation of magnetic protein G beads
The magnetic protein G beads we use are the Dynabeads® protein G from Thermo Fisher Scientific. Protein A beads may be more appropriate to use depending on the species of the derived antibody and its isotype. Also, the use of magnetic beads requires magnetic tube racks that can hold 1.5 ml microcentrifuge tubes for example New England BioLabs #S1506S or other commercially available equivalents.
Prepare the magnetic protein G beads by adding 50 μl of bead volume (1.5 mg of Dynabeads®) to 1.5 ml microcentrifuge tubes, place the tubes with beads in the magnetic rack to separate the beads, and then remove the supernatant with a pipette. Resuspend the magnetic protein G beads in 200 μl of NT2 buffer, and then add 12 μg of the anti-RBP or non-specific antibody. Mix the tubes constantly by rotating end over end for 20 min at room temperature or overnight (~12-18 h) at 4 °C.
After mixing the magnetic protein G beads and antibodies, put the tubes containing the mixture in the magnetic rack to separate the bead-antibody complexes from the buffer. Remove the supernatant, place the tube with the bead-antibody complexes into a regular rack, and then add 500 μl of NT2 buffer. Completely resuspend the bead-antibody complexes by repeatedly inverting the tubes several times and then place them back on the magnetic tube rack. Do this procedure at least three times to wash the beads, and then resuspend the beads in 500 μl of NT2 buffer. Rotate the tubes at 4°C until ready to use.
2.4. Digestion optimization using micrococcal nuclease
It is crucial that nuclease conditions are optimized for each RBP of interest and cell line or tissue. We have optimized conditions for using micrococcal nuclease (MNase). We chose MNase because it requires Ca2+ for activity [28] and is therefore inactivated by chelation of Ca2+ with EGTA. Thus, MNase activity is easily controlled and allows one to optimize conditions that will partially digest RNA. We recommend testing a wide range in the amounts of micrococcal nuclease used, as described below, to ensure that an RNA digestion profile can be observed after electrophoresis.
Prepare lysates from six 15-cm dishes of cells according to the instructions in sub-section 2.2. Begin the digestion optimization experiment by thawing cell lysates on ice and then centrifuging them at 15,000 g, 4 °C for 15 min. This should produce a colloidal pellet at the bottom, and a cleared supernatant. Transfer this cleared supernatant to a clean 1.5-ml microcentrifuge tube. If the supernatant appears cloudy, re-centrifuge at 15,000 g, 4 °C for 5 min, and transfer the cleared supernatant to a clean 1.5-ml microcentrifuge tube. Use one-tenth of the cleared lysate to extract total RNA using TRIsure (Bioline) or TRIzol (Thermo Fisher Scientific) reagent protocols. Resuspend the total RNA in volumes of distilled, nuclease-free water equivalent to the volume of lysate used for total RNA extraction. Measure the total RNA concentration using a spectrophotometer (like a NanoDrop for example), and record the concentration for future reference. Typically, the total RNA concentrations range from 400 ng/μl to 800 ng/μl. The total protein content of the cleared lysate may also be measured using standard Bradford assays. However, total protein is less informative for optimizing nuclease digestions. We have found cleared lysates from HEK293 prepared as described to typically have a total protein concentration ranging from 10-15 μg/μl.
The MNase we use is from New England BioLabs (#M0247S), and is supplied in gel units per ml. The optimal amount of MNase can widely vary depending on cell origin of lysates, RBP studied, antibody used and even person performing the experiments. Because of these variables we strongly recommend optimizing the digestion for each set of experiments. A typical digestion optimization with micrococcal nuclease would be as follows:
○ Mix 300 μl of cleared cell lysate with 0, 125, 500, 2000, 8000, or 32000 gel units MNase.
○ Incubate the reactions at 30 °C for 5 min.
○ Within the final 20 s of the incubation inactivate the MNase by adding EGTA to the reactions to a final concentration of 10 mM, mix by repeated inversion 5 times, then place the digested lysates on ice.
Next, perform immunoprecipitations with the anti-RBP antibodies coupled to magnetic protein G beads. A typical reaction is as follows:
○ For each 500 μl bead-antibody complexes in NT2 buffer, split them evenly in two volumes of 250 μl.
○ Add to each bead-antibody complex mixture 747.8 μl NT2 buffer, 1.2 μl of 1 M DTT, 3 μl of 40 units/μl RNaseOUT, 48 μl of 0.5 M EDTA pH 8.0, and finally 150 μl MNase treated cleared cell lysate. Perform immunoprecipitation by constantly mixing end over end at 4 °C for 2 to 4 h.
○ After immunoprecipitation, place the tubes on the magnetic tube rack to separate the magnetic bead-antibody complexes, remove and save the supernatant, remove the tubes with the bead-antibody complexes from the tube rack, and then add 750 μl of cold NT2 buffer to wash the beads. The saved supernatant represents input lysate after IP, and can be used to compare the efficiency of the IP by western blot. Resuspend the bead-antibody complexes completely by repeated inversions with occasional short pulses using a vortex. Place the tubes back on ice for 10 s and then back in the magnetic tube rack to separate the antibody-bead complexes. Remove and discard the wash-supernatants from the IP, the bead-antibody complexes from the magnetic rack, and then repeat the washes with NT2 buffer for a total of 4 washes.
○ Before discarding the supernatant of the final wash, transfer 37.5 μl (5%) of the resuspended bead-antibody complexes to a clean 1.5 ml microcentrifuge tube for western blot analysis if necessary. Discard the final wash supernatant and then resuspend the bead-antibody complexes in 1 ml of TRIsure (or TRIzol) reagent. Follow the manufacturer's protocol to extract the RNA fragments. RNA fragments can be stored in 8 μl of nuclease-free water at −80 °C if necessary.
○ (Optional, see Section 3.3) If using an input sample, RNA fragments from the input should be depleted of ribosomal RNAs (rRNA). There are several kits commercially available for rRNA depletion. We use Illumina's Ribo-Zero Gold (Human/Mouse/Rat) kit and their protocols to deplete total RNA fragments extracted from the input. 1 μg of the input total RNA fragments is sufficient for the rRNA depletion. The remainder of the input total RNA fragments should be stored at −80 °C.
The RNA fragments must be dephosphorylated and then radiolabeled with [γ-P32]-ATP in order to visualize the MNase digestion products by PAGE through a 15% TBE-Urea polyacrylamide gel. Dephosphorylation is performed using recombinant shrimp alkaline phosphatase (rSAP, New England BioLabs), and radiolabeling is performed using T4 polynucleotide kinase (T4 PNK, New England BioLabs). Typical dephosphorylation and radiolabeling reactions are as follows:
| rSAP Reaction | 1 Reaction |
|---|---|
| Nuclease-free water | 8 μl |
| 10X CutSmart Buffer | 1 μl |
| rSAP (1 U/μl) | 1 μl |
Final reaction volume is 10 μl. If RNA fragments were already stored in 8 μl of water, only add 10X CutSmart Buffer and rSAP. Add 10 μl of the reaction mix to each dried RNA pellet. Incubate the reactions at 37 °C for 30-60 min. Heat inactivate the rSAP by incubating the reactions at 65 °C for 5 min and then place them on ice.
| T4 PNK Reaction | 1 Reaction |
|---|---|
| Nuclease-free water | 6.5 μl |
| 10X T4 kinase buffer | 2 μl |
| [-P32]ATP (6000 Ci/mmol) | 0.5 μl |
| T4 PNK | 1 μl |
Add 10 μl to each sample (20 μl reaction volume) and then incubate at 37 °C for 30 min. Remove the tubes from the heat source, add 10 nmol of non-radioactive ATP (i.e. 1 μl of 10 mM ATP) to each, and then incubate for 15 min at 37 °C. Prepare Decade™ Markers (Thermo Fisher Scientific) according to the manufacturer's protocol. Add 1 volume (20 μl) of 2X gel loading buffer II (Thermo Fisher Scientific) to each sample and the markers, incubate them all at 95 °C for 30 s. Place the samples on ice immediately for 30 s before loading. Load samples on a 15% polyacrylamide, TBE-urea gel skipping at least one lane after each sample. Run the samples in 0.5X TBE at ~100 V for 1.5 to 2hrs at room temperature (until bromophenol blue dye is about 1cm from the bottom of the gel).
Remove the gel from the cassette and wrap it in plastic wrap, and then expose the gel to a blank phosphor imager screen in a phosphor cassette for 3 min (or longer if necessary). Remove the gel from the cassette and scan the phosphor screen on a Typhoon™ scanner (or an equivalent device). Print an ‘actual size’ image of the gel that is dark enough to display prominent gel features (wells, sides and corners of the gel). Select an MNase condition that would provide a distribution of RNA fragments between 25 and 70 nucleotides using the gel as a reference. We have recently published an image of a digestion optimization in [25]. Calculate the ratio of gel units of MNase to μg of total RNA from the selected condition to use going forward as the optimal digestion condition for your DO-RIP-seq experiments. As mentioned previously, this final optimized value can vary depending on many variables and should be determined for one's own experimental conditions, but as a point of reference we have found optimal conditions to range anywhere from 1 unit MNase per μg total RNA to 50 units/μg.
2.5. DO-RIP-seq
After determining the optimal digestion conditions with MNase you can move on to performing a full DO-RIP-seq experiment which includes the preparation of cell lysates and bead-antibody complexes as described in sub-sections 2.2 and 2.3 respectively. Remember that a full DO-RIP-seq experiment involves an IP with non-specific antibodies (negative IP), so when coupling antibodies to protein G beads make sure to set up both RBP-specific and non-specific antibody coupling reactions. Also prepare sufficient cell lysates for the RBP IP and the negative IP.
On the morning of the experiment, thaw the cell lysates on ice, and clear the lysates by centrifugation as described in sub-section 2.2. Also wash the protein G bead-antibody complexes as described in sub-section 2.3 in preparation for the IP. Measure the total RNA concentration in the cleared lysate by extracting total RNA with the TRIsure (or TRIzol) reagent, and then using a spectrophotometer to measure total RNA concentration, as described in sub-section 2.4. Add the MNase to the lysates according to the optimal ratio of gel units MNase to μg total RNA determined by the digestion optimization experiment. Set aside 5% of the MNase digested lysate for use as an input control. This input control can be useful as an ‘insurance policy’ should you have trouble with the negative IPs (see section 3.3). Perform the IPs with anti-RBP and non-specific antibodies coupled to magnetic beads for 2-4 h at 4 °C in NT2 buffer supplemented with DTT, RNaseOUT, and EDTA as described in sub-section 2.4. After IP, wash the IPs 4 times with 750 μl NT2 buffer. Remember to collect one-tenth of the final wash-resuspension of the IPs for western blot of the RBP if necessary. Separate and remove the final wash from the IPs and then add 1 ml of TRIsure (or TRIzol) to extract the RNA fragments. Also extract the RNA fragments from the input controls. Follow the steps outlined in sub-section 2.4 to dephosphorylate, radiolabel, separate radiolabeled RNA fragments by PAGE, and print an ‘actual size’ image of the gel. Use this image to align the gel so that appropriate sizes can be cut. Sizes to be cut should be from 25 to 70 nucleotides. Use a fresh razor blade for each sample to minimize cross-sample contamination. Place gel slices into a clean, non-stick 1.5 ml microcentrifuge tube (Thermo Fisher Scientific, Ambion, product no. AM12450).
2.5.1. Elute radiolabeled RNA fragments from the gel slices
Add 400 μl of 0.4M NaCl to non-stick 1.7 ml microcentrifuge tubes containing the gel slices, briefly centrifuge to get gel slices down into the tube, and then constantly mix the gels overnight at 4 °C. Remove the samples from 4 °C, centrifuge the samples briefly to pull down the gel and supernatant that may get caught in the tube's cap. Use a 200 μl pipet to transfer the supernatant (contains the eluted radiolabeled RNA fragments) from the gel sample to a new non-stick 1.7 ml microcentrifuge tube. Compare the radioactive counts per minute (cpm) of the supernatant to gel to estimate the proportion of radioactive material (RNA fragments) extracted. The first extraction is usually between 40-60% efficient, but most often being closer to 60% efficient. Keep supernatants on ice. Add 200 μl of 0.4M NaCl to the gel samples, and constantly mix again for 1 h at 4 °C. Collect supernatant as before. This step increases extraction yield to >75%. Remove the samples from 4 °C, centrifuge the samples briefly to pull down the gel and supernatant that may get caught in the tube's cap. Transfer the supernatant with a 200 μl pipet to the corresponding tube with the previous supernatant. Add 15-45 μg (1-3ul) of GlycoBlue/glycogen, 1 ml of 100% ethanol, and then incubate the supernatants at −20 °C for 1 h (or −80 °C for 30 min) to precipitate the RNA fragments. Centrifuge the supernatants at 17,000 g, 4 °C for 25 min to pellet the RNA along with the glycogen. Remove the supernatant, wash the pellet with 1 ml of 75% ethanol, and then centrifuge the samples at 12,000 g, 4 °C for 5 min. Remove the supernatant, and repeat this step. Two ethanol washes are recommended to remove extra salt. This is important for downstream salt sensitive ligations.
2.5.2. First strand cDNA synthesis and final cDNA library preparation for sequencing
We use the NEBNext® small RNA library prep protocol (New England BioLabs) as our preferred method for preparing cDNA libraries for sequencing on Illumina Hi-Seq sequencing platforms. We have made one alteration to this protocol by using a custom 5’ RNA adapter that has five random nucleotides at its 3’ end. The sequence of this custom 5’ RNA adapter is GUUCAGAGUUCUACAGUCCGACGAUCNNNNN, where N is any nucleotide. Therefore the design of this primer will produce a pool of oligonucleotides with random 5-nucleotide sequences at the 3’ end. These unique molecular identifiers (UMI) allows for distinguishing between PCR replicated sequences and unique copies [11, 30].
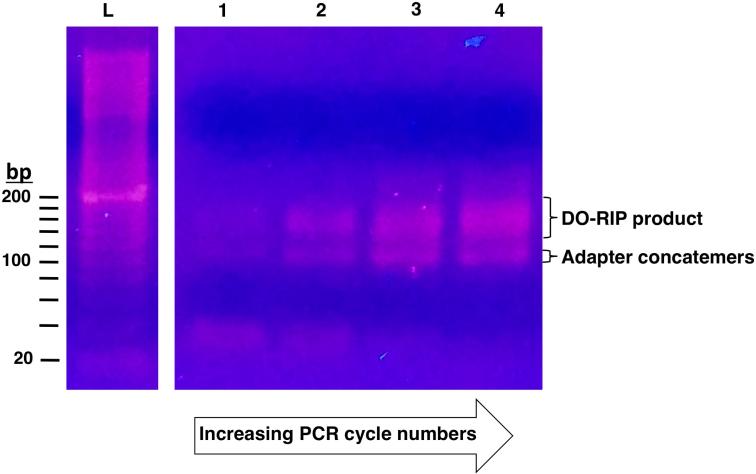
After performing ligations of the 3’ and 5’ adapters to the RNA fragments eluted from the gel, and the reverse transcription to make the first strand of the cDNA, you will need to determine the minimum number of PCR cycles to make a cDNA library for sequencing. The reverse transcription performed using the NEBNext® small RNA library prep protocol is done in a final volume of 40 μl. Prepare a pilot PCR with 4 μl of reverse transcription production in a final reaction volume of 50 μl using the NEBNext® small RNA library prep kit components. Perform the PCR according to the specifications of the NEBNext® small RNA library prep protocol, transferring 4 μl of the reaction to new tubes at various cycle numbers. Cycle numbers that work well are often between 11 and 20, e.g. collect 4 μl of the PCR at cycle numbers 12, 14, 16, 18, and 20. Add 1 μl of 5X DNA sample loading buffer (Bio-Rad) to each of the collected 4 μl fractions and perform agarose gel electrophoresis with a 2.5%, general purpose agarose (Apex™ Bioresearch Products) gel with ethidium bromide, in 1X Tris-acetate-EDTA (1X TAE, 40 mM Tris ph 7.6, 20 mM acetic acid, 1mM EDTA) running buffer. A typical result is showed in figure 2. The cycle number at which a faint amplified product is detected should be used as the number of cycles of PCR to make the final cDNA library for sequencing. Make the final cDNA library using 16 μl of the first strand cDNA from the reverse transcription reactions by performing PCR (100 μl reaction) for the pre-determined number of cycles using the NEBNext® small RNA library prep kit components. Be sure to use the index primers provided with the kit to distinguish cDNA libraries made from the IP and input samples.
Figure 2. Preparing DO-RIP-seq cDNA libraries for deep sequencing.
Ethidium bromide stained, agarose gel showing the cDNA fragments after increasing numbers of PCR cycles (lanes 1-4). Demarcated is the desired product from DO-RIP and below it is the adapter concatemers which must be avoided. Lane “L” is the 20 bp molecular ruler.
2.5.3. cDNA library clean up
As shown in figure 2, the PCR will produce a smear representing the size distribution of the RNA fragments converted into cDNA, and a distinct band below 120 bp, which represents adapter concatamers. These adapter concatemers are the result of ligations of the 3’ and 5’ adapters to each other, and these being made into cDNA. To remove the concatamers we recommend performing a gel purification as follows.
Add 18ul of 5M NaCl (0.3M final concentration) to the completed PCRs, and bring the volume to 300ul using nuclease-free water. Add 300 μl (1 volume) of basic (pH 8.05) phenol chloroform isoamyl alcohol (25:24:1, Thermo Fisher Scientific) to each, vortex for 10 s to mix, and then centrifuge for 15 min at 15,000 g, room temperature (or 4 °C if preferred). Transfer the upper aqueous layer to new tubes then add 1 μl (15 μg) of GlycoBlue™/glycogen and 2.5 volumes of 100% ethanol. Precipitate the DNA by incubating the samples at −20 °C for at least 1 h or −80 °C for 30 min, and then centrifuging at 17,000 g for at least 10 min at room temperature (or 4 °C if preferred). Remove and discard the supernatant, and then wash the cDNA pellet with 1 ml of 80% ethanol. Centrifuge the samples at 9,000 g for 5 min at room temperature (or 4 °C if preferred), and then remove and discard the supernatant. Allow the pellets to air-dry, and then add 16 μl of 1X TAE buffer to dissolve the cDNA pellets.
Prepare a 4% low melt sieve agarose (Apex Bioresearch Products) gel with 1X TAE buffer and ethidium bromide to perform electrophoresis. Use of a 1.5/1.0 mm, 6-tooth comb with the agarose gel is recommended. Add 4 μl of 5X DNA sample loading buffer to the 16 μl cDNA samples, and then load the samples into the wells of the agarose gel (20 μl of cDNA sample per well). Also load 7 μl of EZ Load™ 20 bp molecular ruler (Bio-Rad). Perform electrophoresis at 100 V; low melt agarose gels are easier to melt. Visualize the DNA by exposing the gel to UV light, and then use a clean razor to excise the cDNA product, which should be a smear from approximately 130 to 200 bp in size. Again a sharp band at ~120 bp is adapter concatamer and should be NOT be excised. Transfer the gel slices with cDNA to a clean 15 ml centrifuge tube, and then extract the cDNA from the gel using spin column based gel extraction protocols like the QIAquick® gel extraction kit (Qiagen). Elute the cDNA with either 40 μl of 10 mM Tris-HCl pH 8.5, or 40 μl of distilled, nuclease-free water. You may check how much cDNA was recovered by using a Bioanalyzer Instrument (Agilent), or by performing agarose gel electrophoresis with 2-4 μl of your cDNA through a 2.5% general purpose agarose gel.
2.5.4. Validation of DO-RIP-seq cDNA libraries
DO-RIP-seq libraries can be validated by ligating a fraction of your cDNA into a pGEM®-T vector (Promega), transforming E. coli with the ligated products, and then plating and sequencing (i.e. Sanger sequencing) the colonies for the presence of the inserted cDNA. A typical ligation reaction is as follows:
| pGEM®-T ligation | 1 Reaction |
|---|---|
| Nuclease-free Water | 1 μl |
| 2X Rapid ligation buffer | 2.5 μl |
| pGEM®-T vector | 0.5 μl |
| DO-RIP-seq cDNA | 0.5 μl |
| T4 DNA ligase | 0.5 μl |
Incubate the reactions at either room temperature for 1 h, or 4 °C overnight. The 2X Rapid ligation buffer, pGEM®-T vector, and T4 DNA ligase are components of the pGEM®-T vector system. 1 μl of the ligation product can be used to transform competent E. coli strains like DH5α (Thermo Fisher Scientific), and then the transformants plated and grown on LB agar-ampicillin plates at 37 °C. Pick at least eight visible colonies for screening by PCR using T7 promoter primers (copies sense strand) and SP6 promoter primers (copies anti-sense strand). Purify the PCR products using spin column protocols like QIAquick PCR purification kits (Qiagen), and use one-tenth of the purified PCR product for agarose gel electrophoresis to detect the insertion of DO-RIP-seq cDNA. Inserted product will be ~300 bp in size. Self-ligation products (no insertion) will be under 200 bp in size. Perform Sanger sequencing on the PCR products containing DO-RIP-seq inserts.
Although the number of colonies sequenced is a very small representation of all sequences in the DO-RIP-seq libraries, comparing the number of sequences that map to mRNA to the number of sequences mapping to other non-coding RNAs like rRNA and tRNA is a good indicator of a successful DO-RIP-seq experiment. A good DO-RIP-seq experiment will have at least 50% of the sequences mapping to mRNA (Table 1). Also this validation step will be helpful in determining how well adapter concatamers were avoided. A high quality DO-RIP-seq cDNA library will ideally have no adapter concatamers, but on occasion could have up to 2 out of 10 sequences being adapter concatamers. The DORIP-seq cDNA libraries are now ready for 100 base pair, single read sequencing on Illumina Hi-Seq 2000/2500 or comparable instruments once their validations are deemed satisfactory.
Table 1. Validation of DO-RIP-seq cDNA libraries prior to deep sequencing.
cDNA sequences in fasta format from HuR DO-RIP experiments were inserted into pGEM-T vectors, used to transform E. coli, and colonies were picked for Sanger sequencing. Sequences are from experiments with optimal MNase digestion conditions and over-digestion conditions. Note that over-digestion conditions yielded mostly rRNA fragments, while optimal conditions produce mostly mRNA fragments.
|
HuR DO-RIP sequences from optimal digestion conditions
|
| >CCT8_CDS_chr21:30,434,842-30,435,033_- |
| TTAGTGAGGCTAAACTCAAAATGGGATCTCCGAAGACTTTGTAA |
| >HSPA5_CDS_chr9:128,000,571-128,000,591_- |
| ACAGGTGACCTGGTACTGCTT |
| >PAIP1_CDS_chr5:43,535,713-43,535,744_- |
| AAAGTTGACAGGATCAGTTTTGGAAGATGCTTGG |
| >SON_CDS_chr21:34,929,588-34,929,612_+ |
| AGTCAGGAGGAGCTACTATAGAAGA |
| >LPGAT1_3UTR_chr1:211919329-211919357_- |
| TTGTTTTCTTTGTTCTTTTTTTGAAGTCA |
| >RAN_3UTR_chr12:131,360,755-131,360,795_+ |
| ATCTTGTTTGTTACTGTCATTCCCATTCCTTTTCGTTTAGA |
| >SS18_3UTR_chr18:23597210-23597238_- |
| TATGTTTTTCCCTTTTTTATGTTGGTTGA |
| >SUB1_3UTR_chr5:32,602,122-32,602,167_+ |
| ATTAAGCTTTACTGTTATAGTAGGTAATATGGTTAGTTTGTAGGGA |
| >DNAJB11_intron_chr3:186,296,085-186,296,130_+ |
| TTTTTGTTTCTTCCAGTTTTATTCAATTCCAAACAGCGCTTTTATG |
| >NCOA7_intron_chr6:126,209,995-126,210,024_+ |
| AGTTTCCGTTTTGTTTTTTGCCTTTCCTCC |
| >chrM:14,678-14,726 |
| ACAACGATGGTTTTTCATATCATTGGTCGTGGTTGTAGTCCGTGCGAGA |
| >PPP6R3_5UTR_chr11:68,287,053-68,287,076_+ |
| ACCTGTAATGGGCAAGGAGCCACA |
| >DNAJC14_anti-sense_chr12:56,216,465-56,216,505_+ |
| ACACTCAGCACAGTATCTGGCACTCTTAGGTTCCCGGTCCA |
|
HuR DO-RIP sequences from over-digested conditions |
| >rRNA_chrUn_gl000220:115,785-115,844_- |
| GCAACGGAGGCCATCGCCCGTCCCTTCGGAACGGCGCTCGCCCATCTCTCAGGACCGACT |
| >rRNA_chr17:33,478,208-33,478,234_- |
| ATATTAGTCAGCGGAGGAAAAGAAACT |
| >rRNA_chr21:9,827,183-9,827,220_- |
| GCACGCATCCCCCCCGCGAAGGGGGTCAGCGCCCGTCG |
| >rRNA_chrUn_gl000220:110,738-110,773_- |
| GTACAAAGGGCAGGGACTTAATCAACGCAAGCTTAT |
| >rRNA_chr12:20,704,449-20,704,484_- |
| AATTCCGATAACGAACGAGACTCTGGCATGCTAACT |
| >rRNA_chrUn_gl000220:109,717-109,755_- |
| AATAGCGTATATTAAAGTTGCTGCAGTTAAAAAGCTCGT |
| >rRNA_chr17:33,478,235-33,478,275_- |
| CGCGACCTCAGATCAGACGTGGCGACCCGCTGAATTTAAGC |
| >7SK_chr6:52,860,434-52,860,469_- |
| CCCTGGCGATCAATGGGGTGACAGATGTCGCAGCCA |
| >rRNA_chrUn_gl000220:110,523-110,559_- |
| GCTCAATCTCGGGTGGCTGAACGCCACTTGTCCCTCT |
2.5.5. Processing of raw sequenced reads and data analysis
Raw reads are processed by first removing adapter sequences up to, but not including the five nucleotide UMI using Cutadapt. Secondly, the trimmed reads containing the UMI are collapsed to unique sequences allowing one to distinguish between PCR duplicates and unique copies of a sequence. This step greatly increases sequence yield. Thirdly, the UMIs are removed from the RNA target sequences, and the sequences are mapped to the human genome and transcriptome (or another appropriate genome and transcriptome) using TopHat2 (alternatively, other RNAseq aligners like STAR may be used). In this alignment step it is strongly suggested to use an alignment strategy that includes mapping to an appropriate (i.e. cell line or cell type matched) transcriptome file for improved splice junction mapping and exonic coverage. The uniquely mapped reads can be used to perform two types of analyses; 1) a RIP-Seq-Like (RSL) analysis to measure whole-transcript association, 2) and a binding site (BSL) analysis to quantitatively detect target sites.
RSL analysis is done by first calculating the reads per million (RPM) for each known gene using the Gencode v19 annotations, limited to exonic reads. This analysis is applied to the RBP and negative (and/or input) libraries. RSL scores that quantitatively describe whole transcript association are produced for each gene by calculating the logarithmic difference between each gene's RPM in the RBP and negative (or input) libraries. (Note that the log of a ratio of two numbers is the difference of the logs, so in this case you take the difference of the log transformed RPMs to get the ratio.
In order to normalize the data, BSL analysis is achieved by binning reads from both the RBP library and the negative (or input) library in 5 nucleotide (nt) intervals across the genome used in the mapping step. A pseudo-count of one read is added to intervals with zero reads in instances where the matched library (either RBP or negative) contains non-zero reads. Next, binned read intervals are normalized for library size (per million reads) and log2 transformed. In the case where replicate IPs were performed, reproducible enrichment of reads in the RBP interval are calculated using single-tailed paired t-test and the resulting p-value will be used in the final step of defining the binding site. To limit the influence of stochastic mapping noise a high pass filter is applied where an interval must have at least one sample (RBP or negative for any replicate) with greater than six reads. In cases of low sequencing depth this filter cutoff may need to be decreased. Then, the ratio of normalized RBP reads to normalized negative reads for each 5 nt interval is calculated (ie. the difference of the logs). In the case of replicates, the mean enrichment ratio across the replicates is calculated. Histograms of this log ratio should produce a characteristic bimodal pattern of enrichment, with the two apparent distributions consisting of intervals enriched or depleted in the RBP versus negative. To determine the probabilities of each interval belonging to the RBP enriched distribution mixture models were generated using the R package: ‘mixtools’ as previously described [4]. LOD scores were calculated as the log10 ratio of the posterior probabilities generated from the mixture model. Since it would be expected that an RBP “footprint” from a partial digestion should be >10 nucleotides, we defined binding sites as three or more consecutive 5 nucleotide intervals with LOD > 0 and p < 0.05 (if replicates were used).
As an alternative approach for DO-RIP-seq binding site identification it may be possible to adapt peak calling methods designed for RIP-seq or CLIP to work with DO-RIP-seq data, however we have not tested these alternatives. If attempting an alternative approach it is important to select a method that determines local, strand specific enrichment of RBP binding relative to an empirically measured background, such as ASPeak [31] or RIPSeeker [32].
3. Discussion
3.1. Considerations when selecting a nuclease/ribonuclease for DO-RIP-seq
There are many commercially available nucleases and ribonucleases (RNases) that could be used to partially digest bound RNAs. Micrococcal nuclease was selected because its activity is dependent on calcium ions, and so can be inactivated using EGTA to chelate the calcium ions. However, many other nucleases require divalent cations for activity and thus in theory can be used in DO-RIP-seq. One thing to keep in mind is that some RBPs require divalent cations to bind RNA and so inactivation of the nuclease by chelation would abrogate the RNA binding activity of these RBPs. Competitive inhibitors like vanadyl ribonucleosides or thymidine-3’,5’-diphosphate would be appropriate in this scenario.
In addition the activity of nucleases and RNases can be affected by sequence and secondary structure. Micrococcal nuclease can cut at any nucleotide but has the greatest efficiency for adenosines. RNase T1 for example is specific for guanosine. The nuclease/RNase specificity highlights the importance of optimizing the digestion conditions to prevent significant depletions of certain sequences due to over-digestion.
3.2. Recommendations for performing DO-RIP-seq in different cell lines and RBPs
We have witnessed that the optimized digestion conditions for an RBP in one cell type is different when working with the same RBP in another cell type [26]. Therefore it is important that digestion optimization experiments be performed when working with a new cell line or tissue. We have also observed improvements in cell lysis when the divalent salts (CaCl2 and MgCl2) are left out of the polysome lysis buffer. However, these salts must be added to the lysates before performing the nuclease/RNase digestion of the cell lysate.
3.3. Antibody selection
We recommend using antibodies raised against native proteins; however, DO-RIP-seq may also be preformed using exogenously expressed tagged proteins. In selecting an appropriate antibody it is necessary to verify the specificity of the antibody as well as the suitability for immunoprecipitation. By Western Blotting the antibody should show a single band of the expected size in the input lane, one can also observe the approximate efficiency of the IP by loading lanes with IP, supernatant and negative IP. It is also suggested that when working with a new antibody to confirm that the band of interest is indeed the intended protein; this can be tested by noting whether the band diminishes or disappears in response to siRNA knockdown or genetic knockout.
In the case that specific antibodies are not available, tagged proteins may be used. However there are several additional factors one must consider. First, does the addition of the tag interfere with the function, localization or ability of the RBP to interact with RNA? If it proves to be a problem, moving the tag to the other end of the protein may help. Secondly, overexpressing a RBP that regulates RNA abundance or isoform usage will alter which RNAs are present and approximate the quantity. In this case it is recommended to test a negative sample that also overexpresses the RBP of interest in order to account for any changes in RNA abundance caused by overexpression. For the negative IP one may either use a tagless version of the RBP (as a control for tagged IPs) or use a non-specific antibody, such as a species-matched normal sera or pre-immune sera. Finally, overexpression of an RBP in the presence of the endogenous version of the RBP may result in competition with target sequences and mRNAs. This may cause difficulties in producing quantitative binding values, however this relationship has not yet been explored using DO-RIP-seq so one should proceed with caution.
Finally, if the negative IP lane from the autoradiograph appears blank relative to the positive IP or does not produce enough sequencing coverage for sufficient normalization (i.e. if your negative IP was ‘too clean’), it is possible to construct a control library from the input RNA that was set aside in section 2.5. In our experience, negative IP libraries and input libraries are highly similar and produce nearly identical results. If one chooses to use an input control as an alternative to a negative IP or to replace an insufficient negative IP the protocol is nearly identical except that the IP step is replaced with a rRNA removal step. We have had success using the Ribo-Zero Gold (Human/Mouse/Rat) rRNA removal kit (Illumina, cat. No. MRZG12324), following the manufacturer's standard protocol.
3.4. Replicates
We strongly recommend performing DO-RIP-seq experiments with at least three replicates if possible. Lysates for replicate IPs should be collected at distinct times. For tissue culture cells this ideally means cells plated on separate days. Pooled lysates should never be used as replicates but are recommended for use in positive IP/negative IP pairs.
3.5. Validating targets identified by DO-RIP-seq and grouping targets by function
DO-RIP-seq experiments will yield quantitative metrics of whole transcript targets (RSL scores) and binding sites (LOD scores) of the RBP. Whole transcripts and binding sites with greater RSL and LOD scores are interpreted as being more strongly associated to the RBP. Binding sites with different LOD scores for genes of interest can be biochemically validated using REMSA (RNA electrophoretic mobility shift assays) to measure relative differences in affinity. In a recent study, this assay was performed with HuR DO-RIP-seq binding sites in β-actin, and yielded good correlation with binding site LOD scores [25].
Performing undigested RIP followed by quantitative real time PCR (rtPCR) using primers specific to the genes of interest can validate the whole transcript targets. We have found strong correlations between RIP-rtPCR and RSL enrichment scores from HuR DORIP-seq, and thus, it is a reasonable means to validate the differences in enrichment scores [25]. It is important to note that this protocol is performed without covalent crosslinking, and thus, preserves the natural, non-covalent properties that are present in protein-RNA interactions. Therefore, DO-RIP-seq advances our ability to detect the dynamic changes inherent to a functional post-transcriptional outcome.
3.6. Interpretation of RSL scores and LOD scores
DO-RIP-seq data can be analyzed for RBP-RNA interaction at two different levels: a) whole transcript association (or RIP-seq Like analysis, RSL) and b) binding site association (or Binding Site Level analysis, BSL). The RSL analysis provides quantitative enrichment at the whole transcript level in an IP in proportion to the relative abundance of the transcript in the sample. The RSL analysis is useful for identifying which transcripts are targets and can be used in downstream analyses that make use of gene names for example, gene set enrichment analyses (GSEA, [33]). For example, RSL can be useful when looking for enrichments of groups of functionally related messages (i.e. RNA regulons) or when comparing binding data to transcriptomic measurements, such as RNA abundance or ribosomal profiles. On the other hand, BSL analysis provides quantitative enrichment of local binding sites over relative local background levels. The BSL analysis is useful for identifying sequence or location-based relationships, such as consensus motifs, distances to RNA features, or percentages of sites in transcript regions (i.e. 3’UTRs or introns). These analyses are distinct, but related, and provide complementary information about RBP-RNA association. In essence the RSL analysis can be considered as summing up all the binding site information on a per-transcript basis to deliver quantitative enrichment of transcript-RBP association rather than a discrete yes/no binding determination. The advantage of these twin analyses can be seen in the seamless verification of the relationship between HuR's binding proximity to miR seed sites (BSL analysis) and HuR's ability to antagonize AGO-miR function (RSL analysis) [25].
Highlights.
DO-RIP-seq resolves RBP targeting at the whole transcript and binding site levels
RNA binding sites are identified without the use of crosslinking
Binding site quantitation is achieved by normalizing to background or input RNA
Acknowledgements
We thank Jeff Blackinton for his insights and critical reviewing of the manuscript. JDK declares a financial relationship with MBL Inc. and Ribonomics Inc. We also thank Olivier Fedrigo for discussions on Illumina sequencing technology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tenenbaum SA, et al. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci U S A. 2000;97(26):14085–90. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morris AR, Mukherjee N, Keene JD. Systematic analysis of posttranscriptional gene expression. Wiley Interdiscip Rev Syst Biol Med. 2010;2(2):162–80. doi: 10.1002/wsbm.54. [DOI] [PubMed] [Google Scholar]
- 3.Mazan-Mamczarz K, et al. Identification of transformation-related pathways in a breast epithelial cell model using a ribonomics approach. Cancer Res. 2008;68(19):7730–5. doi: 10.1158/0008-5472.CAN-08-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukherjee N, et al. Coordinated posttranscriptional mRNA population dynamics during T-cell activation. Mol Syst Biol. 2009;5:288. doi: 10.1038/msb.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beisang D, et al. Regulation of CUG-binding protein 1 (CUGBP1) binding to target transcripts upon T cell activation. J Biol Chem. 2012;287(2):950–60. doi: 10.1074/jbc.M111.291658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iruarrizaga-Lejarreta M, et al. The RNA-binding protein human antigen R controls global changes in gene expression during Schwann cell development. J Neurosci. 2012;32(14):4944–58. doi: 10.1523/JNEUROSCI.5868-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keene JD, Tenenbaum SA, Carson CC. Methods for isolating and characterizing endogenous mRNA-protein (mRNP) complexes. 2003 United States patent application 6635422, filed 1999.
- 8.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–5. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 9.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456(7221):464–9. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17(7):909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single- nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29(7):607–14. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedersdorf MB, Keene JD. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol. 2014;15(1):R2. doi: 10.1186/gb-2014-15-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keene JD. Posttranscriptional generation of macromolecular complexes. Mol Cell. 2003;12(6):1347–9. doi: 10.1016/s1097-2765(03)00496-9. [DOI] [PubMed] [Google Scholar]
- 15.van der Brug MP, et al. RNA binding activity of the recessive parkinsonism protein DJ- 1 supports involvement in multiple cellular pathways. Proc Natl Acad Sci U S A. 2008;105(29):10244–9. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanford JR, et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19(3):381–94. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kershner AM, Kimble J. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc Natl Acad Sci U S A. 2010;107(8):3936–41. doi: 10.1073/pnas.1000495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jungkamp AC, et al. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol Cell. 2011;44(5):828–40. doi: 10.1016/j.molcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee N, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43(3):327–39. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks L, 3rd, et al. A multiprotein occupancy map of the mRNP on the 3′ end of histone mRNAs. RNA. 2015;21(11):1943–65. doi: 10.1261/rna.053389.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen HT, et al. Drosophila Imp iCLIP identifies an RNA assemblage coordinating F-actin formation. Genome Biol. 2015;16:123. doi: 10.1186/s13059-015-0687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ennajdaoui H, et al. IGF2BP3 Modulates the Interaction of Invasion-Associated Transcripts with RISC. Cell Rep. 2016;15(9):1876–83. doi: 10.1016/j.celrep.2016.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prasad A, et al. The PUF binding landscape in metazoan germ cells. RNA. 2016 doi: 10.1261/rna.055871.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uren PJ, et al. High-throughput analyses of hnRNP H1 dissects its multi-functional aspect. RNA Biol. 2016;13(4):400–11. doi: 10.1080/15476286.2015.1138030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholson CO, Friedersdorf MB, Keene JD. Quantifying RNA binding sites transcriptome-wide using DO-RIP-seq. RNA. 2016 doi: 10.1261/rna.058115.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smola MJ, et al. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci U S A. 2016;113(37):10322–7. doi: 10.1073/pnas.1600008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saris NE, et al. Magnesium. An update on physiological, clinical and analytical aspects. Clin Chim Acta. 2000;294(1-2):1–26. doi: 10.1016/s0009-8981(99)00258-2. [DOI] [PubMed] [Google Scholar]
- 28.Heins JN, et al. Characterization of a nuclease produced by Staphylococcus aureus. J Biol Chem. 1967;242(5):1016–20. [PubMed] [Google Scholar]
- 29.Draper DE. A guide to ions and RNA structure. RNA. 2004;10(3):335–43. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kivioja T, et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods. 2012;9(1):72–4. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 31.Kucukural A, et al. ASPeak: an abundance sensitive peak detection algorithm for RIP Seq. Bioinformatics. 2013;29(19):2485–6. doi: 10.1093/bioinformatics/btt428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, et al. RIPSeeker: a statistical package for identifying protein-associated transcripts from RIP-seq experiments. Nucleic Acids Res. 2013;41(8):e94. doi: 10.1093/nar/gkt142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]