Figure 2. Identification of conditionally essential genes.
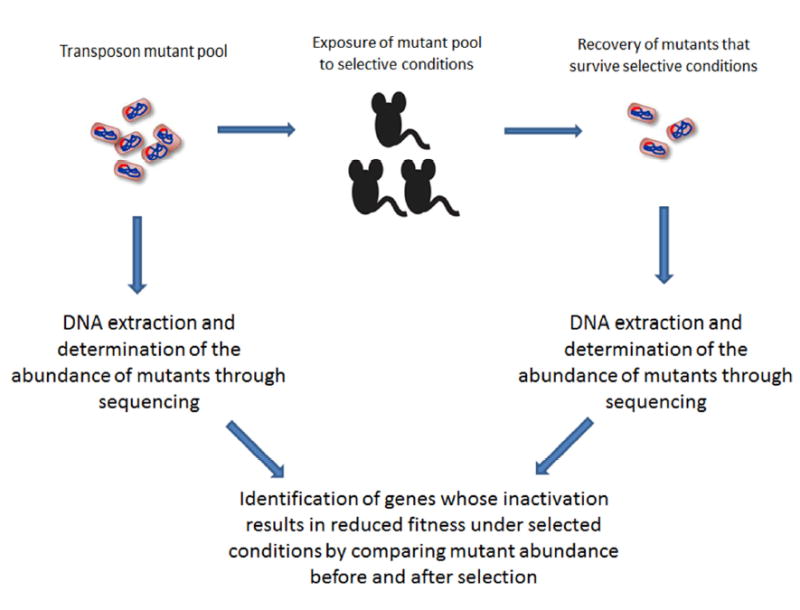
The figure shows a general schematic based on techniques such as HITS, Tn-seq and INseq that employ pools of bacterial transposon mutants to identify genes that contribute to bacterial growth and survival under defined experimental conditions. The pool of transposon mutants is exposed to a selective condition (such as infection in an experimental animal model) and the mutants that survive are recovered. DNA is then extracted from both the original mutant pool and the pool of mutants surviving selective conditions and subjected to next generation sequencing in order to determine the abundance of mutants in each gene present in the pools. The abundance of mutants is compared between pools, and genes that contribute to growth and survival are identified based on the idea that mutants in these genes will demonstrate decreased abundance after selection.
