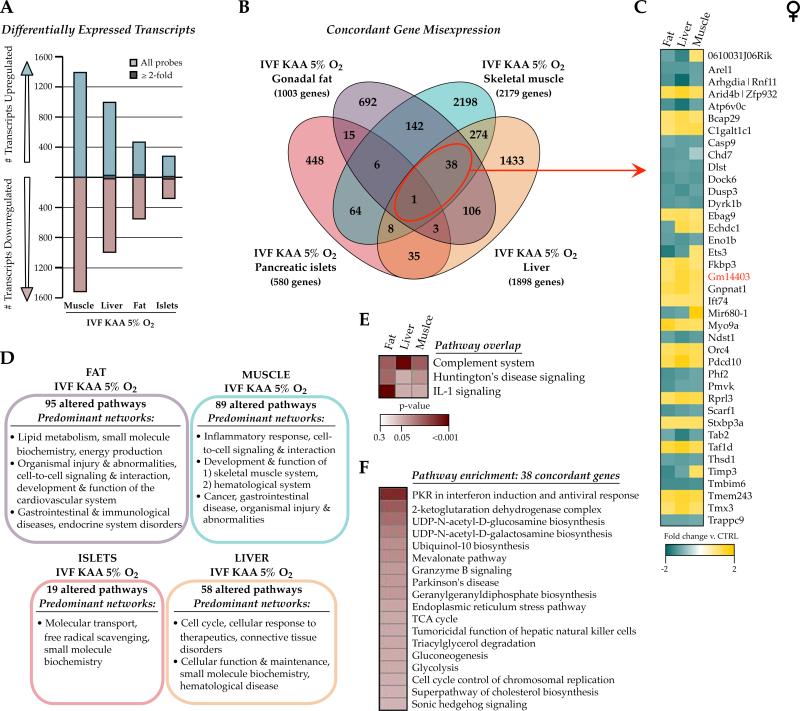
Figure 4. Adult IVF transcriptional signatures are tissue-specific.
Microarray comparison between female 29wk gonadal fat, liver, skeletal muscle and pancreatic islets derived from IVF KAA 5% oxygen animals and in vivo flushed blastocyst controls. A) Number of transcripts altered in each IVF tissue. The majority of changes was modest and ≤2-fold different from controls. B) Venn diagram comparing the concordance of gene misexpression after IVF across the 4 tissues. C) Fold-change directionality of concordant IVF gene misexpression between fat, liver and muscle, with the one gene altered in all 4 tissues (Gm14403) indicated in red. D) Ingenuity Pathway Analysis (IPA) highlighting the predominant networks (network score >30) associated with the gene expression changes in each tissue, and E) the commonly altered pathways between IVF fat, liver and muscle gene lists. F) Top canonical pathways enriched in the 38 genes misexpressed in IVF fat, liver and muscle.