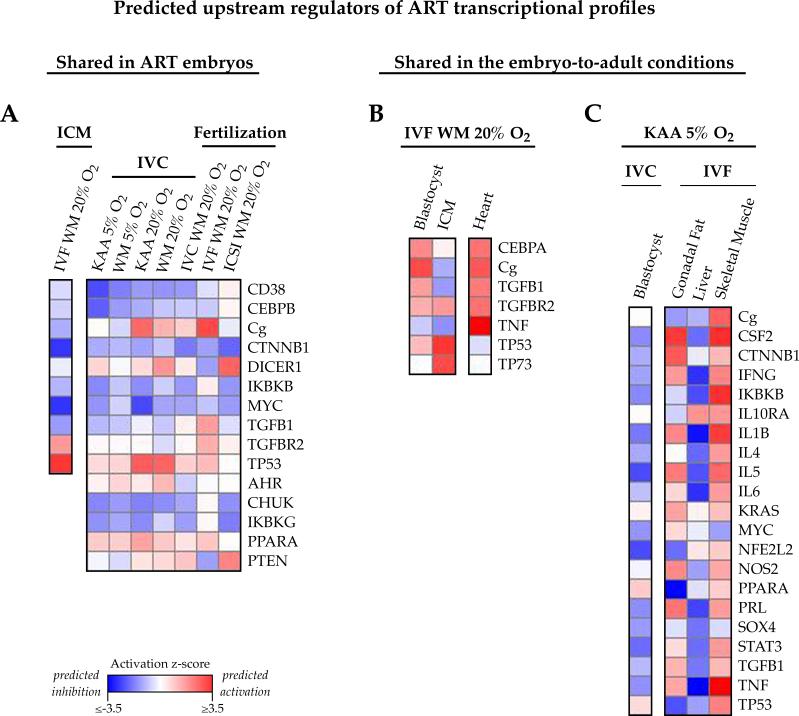
Figure 7. Common upstream regulators may govern the transcriptional changes present in in vitro embryos and adult tissues.
Ingenuity Pathway Analysis identifies regulators functioning upstream of the altered genes, and predicts whether these regulators are impaired (negative z-score, blue) or activated (positive z-score, red) based on fold-change misexpression. A) Shared upstream regulators of the ART embryo expression profiles. 15 molecules were predicted to function upstream of the transcriptional alterations present in the in vitro blastocyst, 10 of which were similarly highlighted in ICM. B, C) Upstream regulators present in the embryo-to-adult condition-specific comparisons. B) 7 regulators commonly target the altered genes in IVF WM 20% blastocysts, ICM and adult heart, and C) 21 regulators collectively mediate the gene expression changes observed in KAA 5% oxygen blastocysts and corresponding adult female fat, liver and muscle.