FIGURE 1.
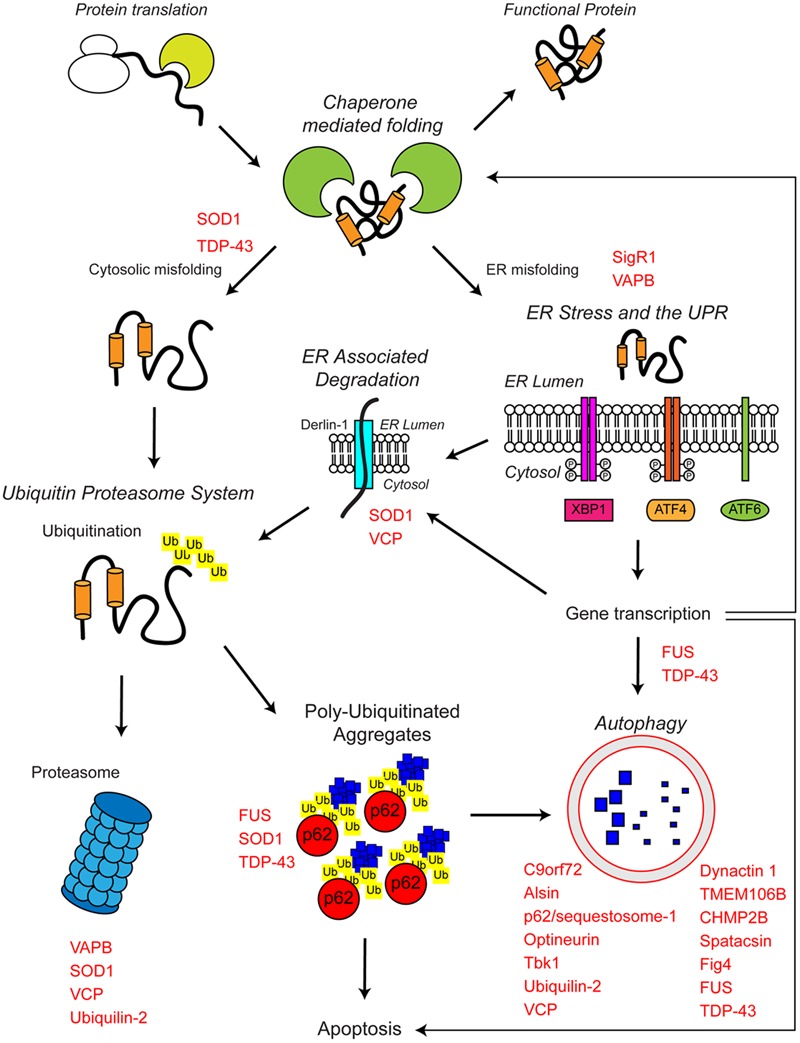
The proteostasis network and ALS. Protein folding occurs co-translationally at the ribosome with the aid of molecular chaperones, including Hsp70. Correct folding is essential for protein function. Protein folding and refolding continues in the cytosol and the endoplasmic reticulum (ER) lumen. Chronic misfolding in the cytosol leads to targeting of misfolded substrates to the ubiquitin proteasome system (UPS). Poly-ubiquitin chains target substrates for degradation by the proteasome. Overwhelming of the UPS can lead to poly-ubiquitinated aggregate formation, which are cleared by the autophagosome–lysosome pathway. Chronic misfolding in the ER leads to the induction of ER stress and activation of the unfolded protein response (UPR). The UPR leads to altered gene transcription, upregulating ER associated degradation (ERAD) and autophagy. The proteostasis network seeks to restore protein homeostasis, but failure of the pathway leads to the aggregation of potentially toxic species. Disruption of the proteostasis network is prevalent in the pathogenesis of ALS. A large number of ALS-associated genes (indicated in red) directly or indirectly regulate the proteostasis network. In addition, some ALS-associated proteins such as FUS, TDP-43, and SOD1 are also substrates of these pathways. For further details please refer to the main text.
