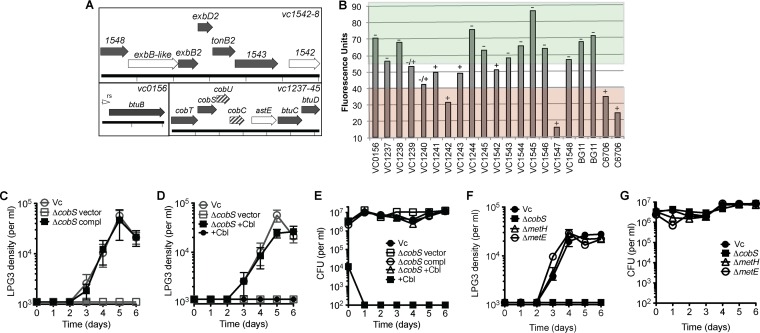
FIG 4.
Corrinoid uptake and remodeling genes are required for V. cholerae support of amoebal growth. (A) V. cholerae genes required (filled arrows), partially required (striped arrows), and not required (open arrows) for supporting amoebal growth on S. elongatus. Cobalamin biosynthesis gene names are assigned using Salmonella nomenclature. vc1241 and vc1243 (flanking astE, not depicted) are not required, and the predicted riboswitch is indicated (rs). (B) Amoeba LPG3 liquid culture screen with V. cholerae transposon mutant library. Cyanobacterial autofluorescence was measured, and LPG3 cell density was scored as follows: −, few or no LPG3 cells; −/+, intermediate number of LPG3 cells; and +, many LPG3 cells. Wells with fluorescence values above 55 did not support amoebal growth (green), wells with fluorescence values below 40 fully supported amoebal growth (red), and wells with intermediate values were scored based on the density of LPG3 cells. Data for two replicates (each) are shown for BG11 and C6706. (C to E) Rescue of V. cholerae cobalamin synthase deletion mutant (vc1238, ΔcobS) for supporting amoebal growth on S. elongatus by genetic complementation (C) and supplementation with cyanocobalamin (Cbl) (D), with corresponding CFU of V. cholerae strains (E). Gray indicates the same growth curve controls. (F) Amoebal growth curves with S. elongatus and V. cholerae mutants of B12-dependent methionine synthase (ΔmetH) and B12-independent methionine synthase (ΔmetE) with corresponding CFU from assay wells (G). Values are averages of triplicate wells plus or minus SEM.