FIG 8.
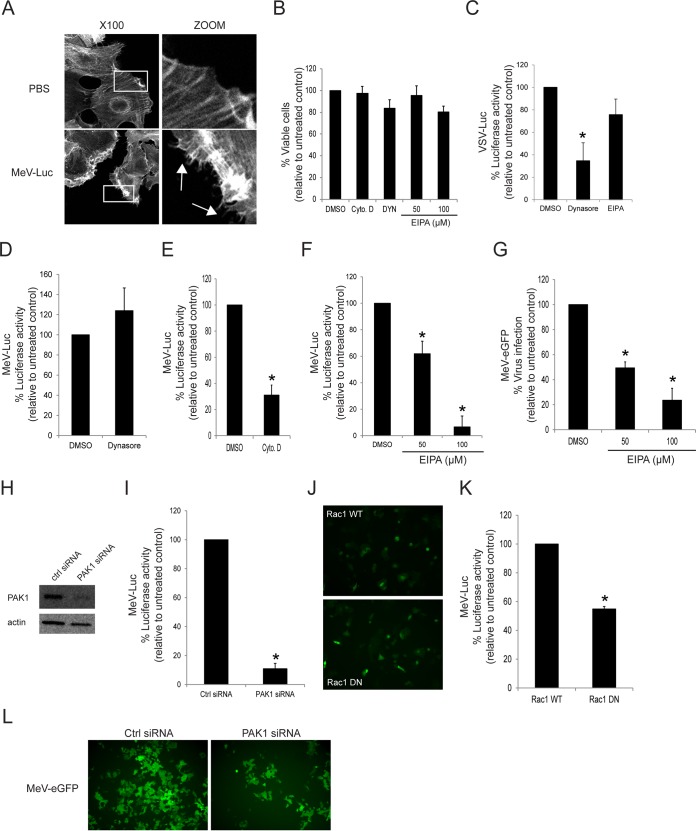
MeV enters the MCF7 human breast cancer cell line through a PVRL4-mediated macropinocytosis pathway. (A) Serum-starved MCF7 cells were exposed to wtMeV-Luc (MOI of 10) or PBS for 30 min and fixed with formaldehyde. Actin filaments were labeled with Alexa Fluor 546-conjugated phalloidin (white). Images were captured with a 100× oil immersion objective. A higher magnification of the boxed area reveals the formation of actin protrusions at the cell surface membrane (arrows). Experiments were repeated 3 times, with similar results. (B) Toxicity assays of MCF7 cells treated with 100 μM Dynasore, 50 and 100 μM EIPA, and 40 μM cytochalasin D were performed by using the MTS cell viability assay, and data are reported as a percentage relative to value for the untreated control (DMSO). (C) As a control, MCF7 cells were pretreated with 100 μM Dynasore or 100 μM EIPA and were infected with VSV-Luc at an MOI of 1. Luciferase activity was measured at 8 hpi, and data are reported as a percentage relative to the value for the DMSO control. Dynasore treatment inhibited the uptake of VSV-Luc, which occurs via the process of CME. However, EIPA treatment had little effect on VSV-Luc entry. (D) MCF7 cells were pretreated with 100 μM Dynasore and infected with wtMeV-Luc. Luciferase activity was measured at 8 hpi, and values are reported as a percentage of luciferase activity relative to that of the untreated control (DMSO). Dynasore did not prevent the entry of MeV-Luc into MCF7 cells. (E) MCF7 cells were pretreated with 40 μM cytochalasin D and infected with wtMeV-Luc. Luciferase activity was measured at 8 hpi, and data are reported as a percentage of luciferase activity relative to that of the untreated control (DMSO). Cytochalasin D greatly reduced the entry of MeV-Luc into MCF7 cells. (F) MCF7 cells were pretreated with 50 μM and 100 μM EIPA and infected with wtMeV-Luc. Luciferase activity was measured at 8 hpi, and values are reported as a percentage of luciferase activity relative to that of the untreated control (DMSO). EIPA inhibited the entry of MeV-Luc into MCF7 cells. (G) MCF7 cells were pretreated with 50 μM EIPA, 100 μM EIPA, or DMSO and incubated with wtMeV-eGFP (MOI of 10) for 1 h. The virus inoculum was removed and replaced with fresh medium containing 200 μM FIP to prevent secondary infection and syncytium formation. Twenty hours later, virus infectivity was measured by FACS analysis, and values are displayed as a percentage of eGFP-positive cells relative to the value for the untreated control (DMSO). EIPA inhibits MeV-eGFP infection of MCF7 cells. (H) MCF7 cells were transfected with siRNA directed against PAK1 or scrambled control siRNA and incubated for 2 days. Cell lysates were subjected to SDS-PAGE and analyzed by Western immunoblot analysis using antibodies directed against PAK1. The knockdown of PAK1 by PAK1-targeting siRNA was confirmed in MCF7 cells. (I) MCF7 cells were transfected with PAK1 siRNA or scrambled control siRNA and incubated for 2 days. The transfected cells were infected with MeV-Luc at an MOI of 2 for 8 h, and luciferase activity is reported as a percentage relative to that of control siRNA. PAK1 siRNA inhibited the entry of MeV-Luc into MCF7 cells. (J) MCF7 cells were transfected with expression plasmids encoding either Rac1 WT or Rac1 DN. The expression plasmids also contained an eGFP reporter gene. Cells were incubated for 2 days, and the synthesis of Rac1 WT or Rac1 DN was confirmed by fluorescence microscopy. (K) MCF7 cells were transfected with Rac1 WT and Rac1 DN expression plasmids and incubated for 2 days. Transfected cells were infected with MeV-Luc at an MOI of 2 for 8 h, and luciferase activity is reported as a percentage relative to that of the Rac1 WT control. Rac1 DN appeared to inhibit MeV-Luc entry. (L) MCF7 cells were transfected with siRNA directed against PAK1 or scrambled control siRNA. After 2 days, cells were infected with wtMeV-eGFP at an MOI of 10. Syncytium formation was observed at 2 days postinfection by using fluorescence microscopy. PAK1 siRNA clearly inhibited MeV-GFP infection. Experiments were repeated 3 times, with similar results. Data from one representative experiment are shown. In the preceding experiments (B to G, I, and K) means of data from three independent experiments are shown, and error bars indicate standard deviations. Statistically significant differences (P < 0.05 by ANOVA) are indicated by asterisks.