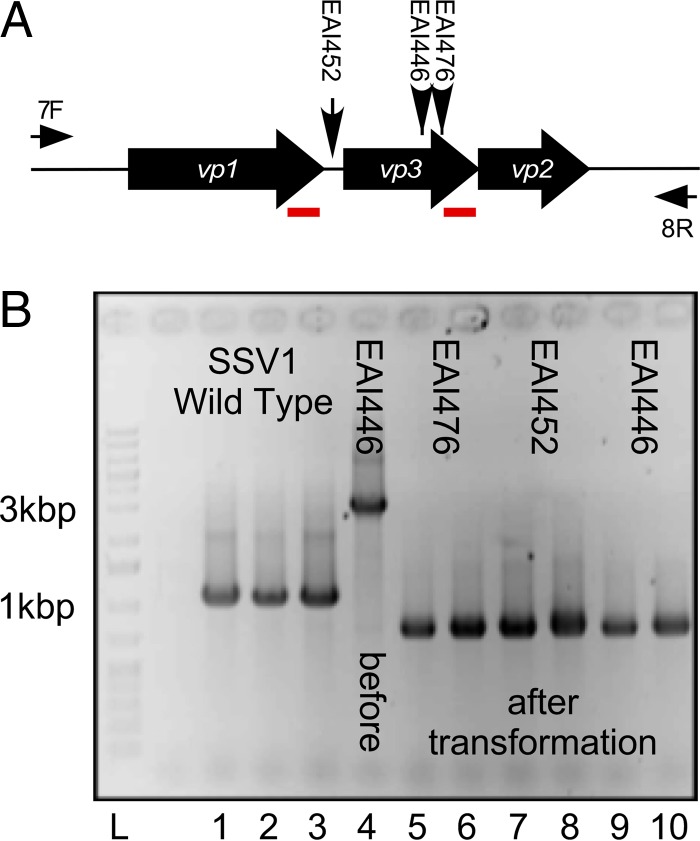
FIG 3.
Insertion mutants in vp3. (A) Overview of vp3 insertion mutants and their analyses. SSV1 structural genes vp1, vp3, and vp2 are indicated as block arrows and labeled in white. The locations of EZ-Tn5 insertion mutants EAI446, EAI452, and EAI476 in the SSV1 vp1/vp3 structural gene region are indicated with vertical arrows. PCR primer (univ_7F and univ_8R) annealing sites are indicated by thin horizontal arrows and labeled 7F and 8R, respectively. Red underlined regions indicate 61-bp direct repeats in the vp1 and vp3 genes. (B) Analysis of vp3 spontaneous deletions. PCR with primers univ_7F and univ_8R was performed on DNA purified from transformed Sulfolobus sp. strain S441 (except lane 4, where DNA was purified from transformed E. coli). Templates: lanes 1 to 3, wild-type SSV1; lane 4, EAI446 DNA purified from E. coli used to transform S441; lanes 5 to 10, DNA from Sulfolobus species transformed with EAI476 (lanes 5 and 6), EAI452 (lanes 7 and 8), and EAI446 (lanes 9 and 10). Lane L, GeneRuler 1-kb plus DNA ladder (Fisher). Relevant molecular masses are indicated beside the gel.