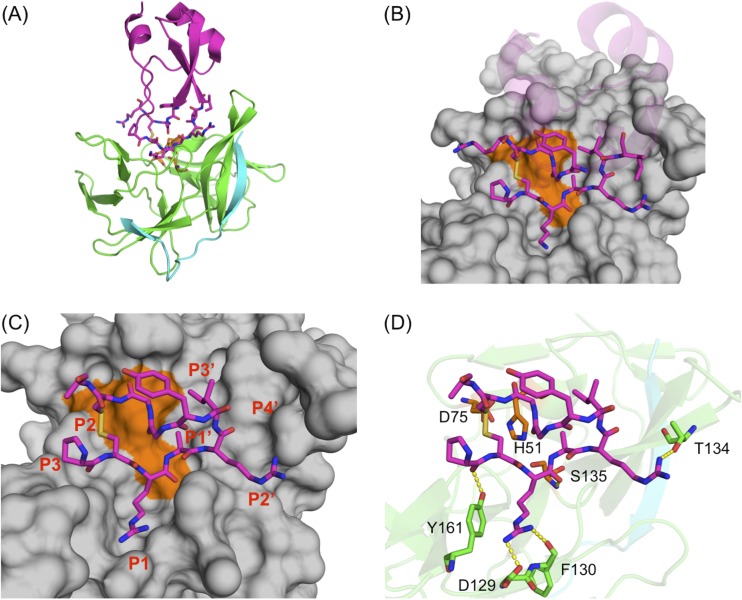
FIG 1.
Design of cyclic peptides derived from aprotinin as dengue virus NS3/2B protease inhibitor. (A) Aprotinin-DENV3 protease complex structure (PDB code 3u1j). The NS3 protease domain is in green, the NS2B cofactor is in cyan, and aprotinin is in purple. (B) The binding loop and the second loop of aprotinin on which the cyclic peptides were based are displayed as sticks. Dengue virus NS3/2B protease is shown as a gray surface, and the catalytic triad is indicated by orange. (C) Cyclic peptide derived from aprotinin is displayed as sticks. The amino acids (P3 to P3′) are numbered based on substrate residue positions. (D) Hydrogen bonds between the cyclic peptide (magenta sticks) and dengue virus NS3/2B protease (residues labeled and side chains depicted as green sticks) are displayed as yellow dashes. The catalytic triad of protease is depicted as orange sticks, and residues are labeled.