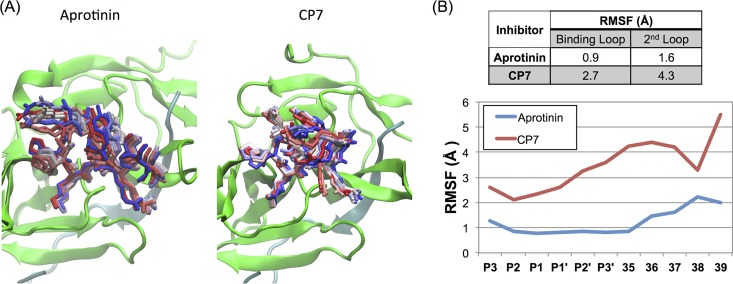
FIG 2.
Molecular dynamics (MD) simulations of aprotinin and the cyclic peptide CP7 bound to the dengue virus protease active site. (A) Snapshots from MD simulation trajectories of aprotinin and CP7 bound to DENV3 protease. The protease is depicted in cartoon representation in green and the aprotinin's binding/second loops or CP7 as sticks from the start (in red) to the end (in blue) of the trajectory. (The simulations were performed with full-length aprotinin, but only residues corresponding to native substrates are shown for clarity.) (B) The RMSF values during MD simulations were tabulated as the average for the binding loop and the second loop and are displayed for individual residue positions.