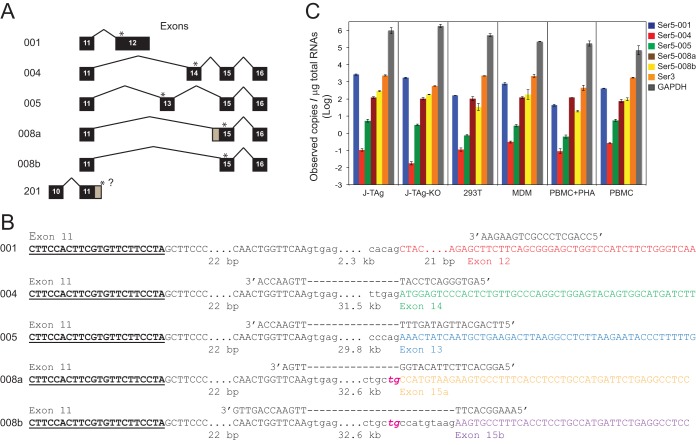
FIG 3.
Detection of human Ser5 gene expression by RT-qPCR. (A) Human Ser5 gene alternatively spliced isoforms. Isoform numbers are shown on the left, and the differentially spliced exons are shown on the right. Exons are shown as black boxes with numerals, but the first 10 exons, which are constitutively spliced, are not shown (except for the putative isoform 201). Exons and introns are not drawn to scale, although exon 12 for isoform 001 is shown as a longer exon. The positions for the stop codons for each isoform are indicated by asterisks. The gray regions for single exons for isoforms 008a and 201 indicate the alternative splice points for the two isoforms. The question mark after exon 11 indicates that the end of the 3′ untranslated region for the isoform has not been identified. (B) Locations of forward and reverse RT-qPCR primers for each Ser5 isoform. The locations of the forward primers (boldface underlined letters) are the same for each primer set. Uppercase letters represent exonic regions, and lowercase letters represent intronic regions. The ellipses indicate sequences that are not shown (the numbers of bases that are not displayed are given below). The dashed lines indicate the intronic regions crossed by the reverse primers. Exons 11, 12, 13, 14, 15a, and 15b are shown in different colors. Instead of the canonical AG normally found for a U2 splice acceptor sequence, 008a and 008b have the rare TG acceptors, which are italicized and shown in magenta. (C) Ser3 and Ser5 copy numbers in J-TAg, J-TAg-KO, and 293T cells; MDMs; PBMCs; and PBMCs activated with PHA. GAPDH was used as a control. The GenBank accession numbers for Ser5-001, -004, -005, -008a, and -008b are NM_001174072.2, BC101280, BC101281, AF498273, and BC101280, respectively. No GenBank accession number was found to be associated with Ser5-201.