Abstract
The H3 lysine 36 histone methyltransferase SETD2 is mutated across a range of human cancers. Although other enzymes can mediate mono- and dimethylation, SETD2 is the exclusive trimethylase. SETD2 associates with the phosphorylated carboxy-terminal domain of RNA polymerase and modifies histones at actively transcribed genes. The functions associated with SETD2 are mediated through multiple effector proteins that bind trimethylated H3K36. These effectors directly mediate multiple chromatin-regulated processes, including RNA splicing, DNA damage repair, and DNA methylation. Although alterations in each of these processes have been associated with SETD2 loss, the relative role of each in the development of cancer is not fully understood. Critical vulnerabilities resulting from SETD2 loss may offer a strategy for potential therapeutics.
SETD2 trimethylates histone H3 at lysine 36, causing effector proteins for various processes (e.g., DNA damage repair) to bind. Mutation or loss of SETD2 may disrupt these processes and lead to cancer.
Large-scale sequencing efforts of human cancers have identified recurrent mutations and deletions in a variety of chromatin regulatory proteins. SETD2, a methyltransferase that trimethylates histone H3 at lysine 36 (H3K36me3), has been found to be both mutated and deleted in select range of cancers suggesting a role in tumor suppression. In this review, we will discuss the range of SETD2 mutations and how disruption of this gene could foster the development of cancer.
ENZYMATIC FUNCTION AND STRUCTURE OF SETD2
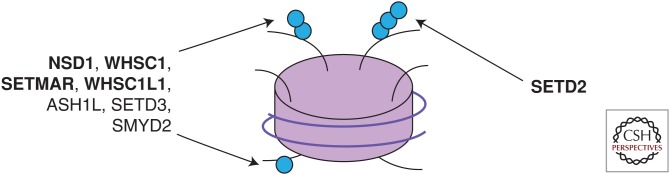
SETD2 (also known as HYPB or KMT3a) is the sole methyltransferase that mediates trimethylation of histone H3.1 and H3 variants (Fig. 1) (Edmunds et al. 2008). Although SETD2 shows biochemical evidence of mono- and dimethylation activity, in cells it seems to exclusively mediate trimethylation because SETD2 silencing results in a near complete loss of H3K36 trimethylation without decreasing mono- or dimethylation levels (Edmunds et al. 2008; Yuan et al. 2009). In higher eukaryotes, other enzymes, including NSD1, WHSC1 (NSD2), and SETMAR, are able to mono- and dimethylate H3K36 (reviewed in Wagner and Carpenter 2012). In contrast, the homolog of SETD2 in Saccharomyces cerevisiae, Set2, exclusively mediates all H3K36 methylation states (Strahl et al. 2002). Although extensive genetic experimentation with yeast Set2 has informed our understanding of the biochemical properties of critical SETD2 domains as well as the possible roles, the redundancy of mono- and dimethylation activities in humans offers an important caveat when extrapolating results from S. cerevisiae.
Figure 1.
H3K36 methyltransferases. Methyltransferases shown to mono-, di-, and trimethylate H3K36. Those shown in bold have been shown in cell-based assays and/or in vivo. (Figure adapted from data in Wagner and Carpenter 2012.)
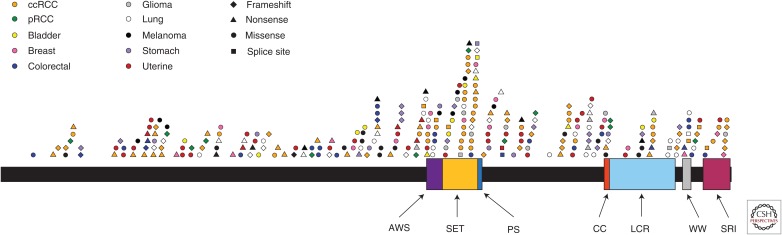
The functions of SETD2 have been attributed to several domains in the protein (Fig. 2). These domains share sequence homology as well as functional similarity with those in yeast Set2. The methyltransferase activity is mediated by the conserved SET domain (Strahl et al. 2002; Li et al. 2005). SETD2 contains two known protein-binding domains: SRI (Set2 Rpd1 Interacting) and WW. The SRI domain mediates association with the hyperphosphorylated carboxy-terminal domain (CTD) of RNA polymerase II (RNAPII) (Li et al. 2005; Sun et al. 2005; Xiao et al. 2003). In yeast, this interaction is required for H3K36 activity, as deletion of the RNAPII CTD decreases H3K36 methylation levels (Li et al. 2003; Xiao et al. 2003). The WW domain, which precedes the SRI domain, may mediate intramolecular interaction (Gao et al. 2014). Based on the property of WW domain interaction with phosphorylated proteins, this domain could also mediate other protein interactions (Lu et al. 1999). About half of SETD2 consists of a large amino terminal domain that is not shared by yeast Set2 and is of unknown function.
Figure 2.
Schematic representation of SETD2 and cancer-associated mutations. SETD2 domains: AWS (associated with SET), SET (Su(var)3-9, enhancer-of-zeste, trithorax), PS (post-SET), CC (coiled coil), LCR (low charge region), WW, SRI (Set2 Rpb1 Interacting). Mutation lists were obtained from CBioPortal on February 5, 2016, and separated into cancer types. Duplicates were removed. Mutations are plotted by color (cancer type) and shape (mutation type). ccRCC, Clear cell renal cell carcinoma; pRCC, papillary renal cell carcinoma.
SETD2 MUTATION IN CANCER
Hundreds of distinct SETD2 mutations have been identified across a wide range of human tumors, including epithelial, central nervous system (CNS), and hematopoietic (Fig. 2; Table 1) (Gao et al. 2013; Cerami et al. 2012). SETD2 mutation was first described in clear cell renal cell carcinoma (ccRCC). In a cohort of 407 ccRCC tumors, truncating mutations were observed in 12 samples (Dalgliesh et al. 2010). SETD2 mutation was also found in ccRCC cell lines (Duns et al. 2012). In The Cancer Genome Atlas (TCGA) study of ccRCC, SETD2 was the third-most-commonly mutated gene with a prevalence of 15.6%. SETD2 is located on chromosome 3p, which shows near universal loss of heterozygosity in ccRCC (Zbar et al. 1987). Chromosome 3p is also the location of the well-known tumor suppressor von Hippel–Lindau (VHL). VHL expression is lost in most cases of sporadic ccRCC, and germline mutation is associated with a high penetrance of ccRCC (Stolle et al. 1998). Mutations of SETD2 affect the remaining allele, and frequently have a significant impact on gene function. Most mutations tend to be inactivating frameshift or nonsense mutations, although missense mutations in critical domains have been detected (Gerlinger et al. 2012; The Cancer Genome Atlas 2013a; Gossage et al. 2014; Simon et al. 2014). In a study of 128 sporadic ccRCC tumors that specifically examined genes known to be mutated in ccRCC, SETD2 was mutated in ∼16% (Gossage et al. 2014). Five frameshift, 10 nonsense, and two splice site mutations were observed. Of three nonsynonymous missense mutations, one altered the SET domain (Gossage et al. 2014). Studies of intratumoral genetic heterogeneity also support a key role for SETD2 loss. Sequencing multiple sites in a single kidney tumor together with metastatic sites identified multiple distinct SETD2 mutations each likely to disrupt function. This convergent tumor evolution suggests that SETD2 mutation is a critical event for a subset of ccRCCs (Gerlinger et al. 2012). Suggestive of a link with aggressive disease, a lower level of H3K36 trimethylation was observed in tumors in metastases compared with primary tumors (Ho et al. 2015).
Table 1.
Cancers associated with SETD2 mutation
| Cancer type | Mutation % (total samples) | References |
|---|---|---|
| Clear cell renal cell carcinoma | 15.6% (418 samples) | The Cancer Genome Atlas 2013a |
| High-grade glioma | 15% (543 samples) | Fontebasso et al. 2013 |
| Uterine carcinosarcoma Uterine corpus endometrioid carcinoma |
13.6% (22 samples) 9% (240 samples) |
Jones et al. 2014 The Cancer Genome Atlas et al. 2013 |
| Acute lymphocytic leukemia | 12% (125 samples) 10% (94 samples) |
Zhang et al. 2012; Mar et al. 2014 |
| Bladder urothelial carcinoma | 10.2% (107 samples) 6% (50 samples) |
The Cancer Genome Atlas 2014b; Van Allen et al. 2014 |
| Desmoplastic melanoma Melanoma Cutaneous melanoma |
10% (20 samples) 8% (25 samples) 5.5% (91 samples) |
Berger et al. 2012 Shain et al. 2015 Berger et al. 2012; Hodis et al. 2012 |
| Lung adenocarcinoma | 9% (230 samples) 5.5% (182 samples) |
Imielinski et al. 2012; The Cancer Genome Atlas 2014c |
| Colorectal adenocarcinoma | 8.3% (72 samples) 6.1% (212 samples) |
The Cancer Genome Atlas 2012; Seshagiri et al. 2012 |
| Pancreatic adenocarcinoma | 8.3% (109 samples) | Witkiewicz et al. 2015 |
| Stomach adenocarcinoma | 7% (287 samples) | The Cancer Genome Atlas 2014a |
| Papillary renal cell carcinoma | 7.6% (157 samples) | The Cancer Genome Atlas et al. 2016 |
| Cutaneous squamous cell carcinoma | 6.9% (29 samples) | Li et al. 2015 |
Cancers are selected for which the mutation rate in CBioPortal (Cerami et al. 2012; Gao et al. 2013) exceeded 5% and a publication was available. Indicated mutation rate reflects published results. Additional cancers discussed in the text are also included.
SETD2 mutations have also been identified in multiple other cancers. High-severity SETD2 mutation was observed in 15%–28% of pediatric and 8% of adult high-grade gliomas (HGGs) (Fontebasso et al. 2013). In contrast, SETD2 mutations were not identified in low-grade gliomas. In addition, all tumors with SETD2 mutation were located in the cerebral hemispheres (Fontebasso et al. 2013). However, mutation of SETD2 was detected in a single diffuse intrinsic pontine glioma, although it co-occurred with an H3.1 K27M mutation, a common feature of these tumors (Wu et al. 2014). Intriguingly, high-grade hemispheric gliomas of children and young adults are also commonly associated with mutation of histone H3.3 (H3F3A) at glycine 34 (Schwartzentruber et al. 2012; Wu et al. 2012). These mutations were nonoverlapping with SETD2 and were associated with reduced H3K36 methylation (Schwartzentruber et al. 2012; Sturm et al. 2012; Fontebasso et al. 2013). Gliomas also commonly harbor IDH1 mutations, which result in the generation of the 2-hydroxyglutarate (2-HG) oncometabolite (Dang et al. 2009). These mutations are not mutually exclusive with SETD2 mutation. Although 2-HG inhibits histone demethylases, including those that can act on H3K36, it is not clear whether IDH1 mutation directly affects H3K36me3 (Chowdhury et al. 2011; Xu et al. 2011; Lu et al. 2012; Fontebasso et al. 2013). Overall, these findings indicate that dysregulation of H3K36me3 is a common event in glioma. The finding linking H3.3 mutation with reduced H3K36 methylation will be discussed in more detail below.
SETD2 mutations have also been identified in acute leukemias. In a study of early T-cell precursor acute lymphoblastic leukemia (ETP-ALL), approximately 10% showed deletion or high-severity mutation of SETD2 (Zhang et al. 2012). In a separate study, SETD2 mutation was detected in approximately 6% of ALL and AML samples (Zhu et al. 2014). Mutation of SETD2 was more common in both acute lymphoid and myeloid leukemias with mixed lineage leukemia (MLL)-rearrangement compared to acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) with an intact MLL gene. Further supporting SETD2 loss as a critical event in leukemia development, SETD2 mutations were commonly nonsense or frameshift, and approximately a quarter of samples carried biallelic SETD2 mutations (Zhu et al. 2014). A comparison of matched primary and relapsed ALL samples suggested that mutations in epigenetic regulators as a class were more common at relapse, and this included mutations in SETD2 (Mar et al. 2014). In this study, SETD2 showed a mutation rate of 5% in a pilot cohort and 12% in a larger validation set. The validation set contained a higher fraction of MLL-rearranged samples possibly explaining the discrepancy in mutation frequency. Suggesting that the importance of SETD2 mutation is greater in acute leukemias in children, the study of AML by TCGA identified only a single SETD2 mutation among 191 adult samples (The Cancer Genome Atlas 2013b). The link between SETD2 loss and MLL rearrangement is provocative since, like SETD2, MLL fusion proteins can associate with components of the transcriptional complex, and the combined alterations in these proteins may lead to transcriptional dysregulation (Milne et al. 2010; Yokoyama et al. 2010).
SETD2 mutations have also been observed at a low frequency in a range of other tumors. 6% of melanoma and chronic lymphocytic leukemia showed SETD2 alteration (Berger et al. 2012; Lee et al. 2015; Parker et al. 2016). SETD2 alterations were observed in high-risk, but not low-risk, gastrointestinal stromal tumors (Huang et al. 2015). SETD2 mutation has also been described in phyllodes tumors of the breast, but not in breast fibroadenoma (Tan et al. 2015; Liu et al. 2016). Among other genitourinary tumors, SETD2 mutation is found in 10% of bladder tumors and the papillary subtype of renal cell carcinoma (The Cancer Genome Atlas 2014b; The Cancer Genome Atlas et al. 2016). Although many of these mutations are monoallelic and consequently predicted to lead to haploinsufficiency, mutations are not the exclusive mechanism for modulating SETD2 activity.
Decreased H3K36me3 has also been observed in the context of nonmutant SETD2 in ccRCC (Simon et al. 2014). miR-106b-5p, a micro RNA known to regulate SETD2, was elevated in a cohort of 40 ccRCC tumor samples; levels of this miRNA inversely correlated with SETD2 mRNA and protein levels (Xiang et al. 2015). SETD2 mRNA levels were also decreased in a subset of patients with AML and lymphoma (Zhu et al. 2014).
In addition to alterations in SETD2, H3K36me3 can be lost by mutation of the methyl acceptor site in histones or by mutations in neighboring amino acids. Virtually all chondroblastomas harbor a lysine 36 to methionine variant in histone H3.3 (H3.3K36M) (Behjati et al. 2013). Expression of the H3.3K36M mutant in cells led to depletion of all H3K36 trimethylation (Lewis et al. 2013). H3.3K36M binds and directly inhibits the activity of SETD2 and the dimethylation activity of MMSET (Fang et al. 2016). Mutations in H3.3 at the neighboring G34 residue have been described in almost all giant-cell bone tumors (Behjati et al. 2013) and, as previously mentioned, in high-grade gliomas (Schwartzentruber et al. 2012; Wu et al. 2014). Overall, the common finding of loss or inhibition of SETD2 across a wide range of cancers suggests the importance of disrupted H3K36 methylation in cancer development.
TRANSCRIPTION AND RNA SPLICING
Chromatin influences many cellular processes including transcription, replication and DNA damage repair. Many studies have linked SETD2 and transcription. In yeast, Set2 and H3K36 trimethylation are associated with gene bodies, and H3K36me3 levels correlate both with the level of transcription and the position in the gene (Krogan et al. 2003; Bannister et al. 2005). H3K36me3 signals are enriched at exons, although the higher levels of nucleosome occupancy at exons may explain this difference (Schwartz et al. 2009). Treatment with a transcription inhibitor causes a decrease in H3K36me3 levels (de Almeida et al. 2011), suggesting that active transcription is necessary for H3K36me3 placement. These data are consistent with the model that Set2 and SETD2 are targeted to elongating RNAPII.
Our understanding of the role of SETD2 in transcription is largely based on studies of Set2 in yeast. In yeast, Set2 partially functions to prevent cryptic initiation, aberrant transcription from internal sites. H3K36 methylation recruits the Rpd3C(S) complex, which includes a histone deacetylase (Carrozza et al. 2005; Keogh et al. 2005), leading to the deacetylation of histones in gene bodies. H3K36 methylation also suppresses of the interaction between histone H3 and the histone chaperone Asf1 (Venkatesh et al. 2012). Preventing the incorporation of new acetylated histones maintains a hypoacetylated state, thereby stabilizing nucleosomes that decreases the chance of a segment of gene being aberrantly recognized by the transcriptional initiation complex as a promoter (Carrozza et al. 2005; Li et al. 2007; Lickwar et al. 2009).
The role of SETD2 in transcription in higher eukaryotes is more complicated. In addition to the separation of methylation activities across multiple enzymes, genes in higher eukaryotes contain introns and are regulated by alternative splicing, and DNA itself can be methylated. H3K36 methylation levels differ based on exon utilization (Kolasinska-Zwierz et al. 2009), with alternatively spliced exons having lower levels of H3K36me3 than those that are constitutively included. Altering SETD2 levels influenced the inclusion of exons in genes known to be alternatively spliced (Luco et al. 2010). Deletion of the splice acceptor site in the β-globin intron leads to shifts in H3K36me3 signal (Kim et al. 2011), and intronless genes have lower levels of H3K36 trimethylation (de Almeida et al. 2011). Chemical or RNAi-mediated inhibition of splicing decreased H3K36me3 levels. Together these data suggest a close relationship between trimethylation of H3K36 and RNA splicing. Perhaps reflecting aberrant transcription or RNA processing, silencing SETD2 results in mRNA accumulation in the nucleus (Yoh et al. 2008).
Several studies have explored the impact of SETD2 loss on transcription in kidney cancer. Examining primary ccRCC, H3K36me3-deficient tumors show alterations in splicing and evidence of intron retention (Simon et al. 2014). This association was also detected in transcriptomic ccRCC data from TCGA. Similarly, differential splicing and altered exon utilization was observed in SETD2 knockout cells (Ho et al. 2015). However, no difference in exon usage or intron retention was observed in other studies in which SETD2 was silenced using RNAi (Kanu et al. 2015). SETD2 has also been associated with aberrant transcriptional termination (Grosso et al. 2015). In the absence of appropriate termination, RNAPII complexes can read through into neighboring genes yielding chimeric RNAs.
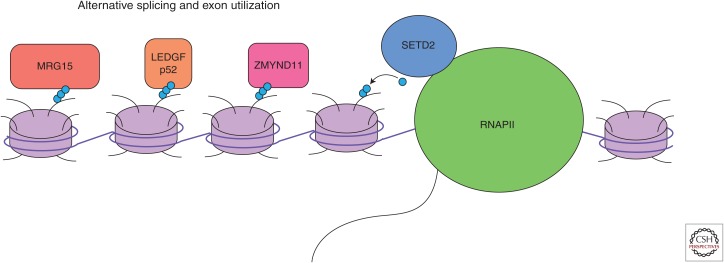
Several proteins that bind H3K36 methylation offer a link between SETD2 and RNA splicing (Fig. 3). LEDGF (PSIP1) exists as two isoforms, p52 and p75, based on the inclusion of six additional 3′ exons (Singh et al. 2000). Both forms contain a PWWP domain, which interacts with di- and trimethylated H3K36. The short form, LEDGF/p52, interacts with proteins involved in alternative splicing (Pradeepa et al. 2012). ZMYND11, also a PWWP domain–containing protein, binds H3.3K36 and associates with regulators of RNA splicing (Guo et al. 2014). MRG15 is a chromodomain-containing protein that binds di- and trimethylated H3K36 (Zhang et al. 2006) and recruits polypyrimidine tract-binding protein (PTB) to alternatively spliced exons (Luco et al. 2010). PTB then binds to silencing elements causing repression of specific exons.
Figure 3.
Transcription and RNA processing. SETD2 associates with RNAPII to posttranslationally modify nucleosomes. H3K36me3 is directly bound by readers, which mediate RNA processing through downstream effectors.
In embryonic stem cells, MRG15 also recruits the lysine demethylase KDM5B to H3K36me3 marked chromatin (Xie et al. 2011). Silencing KDM5B resulted in recruitment of unphosphorylated RNAPII to intragenic regions marked by H3K4me3, potential sites of cryptic initiation. Knockdown of KDM5B increased levels of unspliced transcripts, possibly reflecting aberrant transcription. Downregulation of SETD2 was also found to be associated with increased RNA abundance at noninitiating exons, potentially indicating transcriptional initiation from these sites (Carvalho et al. 2013). Taken together, the extent to which cryptic initiation in yeast is a function of dimethylation (rather than trimethylation) or that RNA alteration in higher eukaryotic cells results from aberrant splicing (rather than cryptic initiation) are unknown.
Overall, SETD2-mediated histone modification and its interaction with specific binding partners offers an explanation for the variation in transcription and aberrant RNA detected after SETD2 loss. Although intron retention has been shown to be a mechanism of tumor-suppressor inactivation (Jung et al. 2015), how SETD2 loss facilitates tumor development remains unknown.
CHROMATIN STRUCTURE
Several studies have shown that H3K36me3 loss results in alterations in chromatin architecture. Silencing SETD2 impairs the recruitment of the Facilitates Chromatin Transcription (FACT) complex to transcribed chromatin, which results in increased sensitivity to MNase digestion, suggesting alteration in nucleosomal interaction with DNA (Carvalho et al. 2013). This effect was particularly evident at internal exons. Chromatin alterations in response to SETD2 silencing was also observed in a kidney cancer cell line (Kanu et al. 2015). A similar observation was made by examining chromatin accessibility in H3K36me3-deficient primary renal cell carcinomas using formaldehyde-assisted isolation of regulatory elements (FAIRE) (Simon et al. 2014). Overall, enhanced accessibility corresponded to regions typically marked by H3K36me3. By examining individual genic features, signal increases were most striking immediately preceding exons, suggestive of a specific effect at splice acceptor sites. Together, these studies support a link between SETD2, chromatin accessibility and splicing.
DNA REPLICATION AND DAMAGE REPAIR
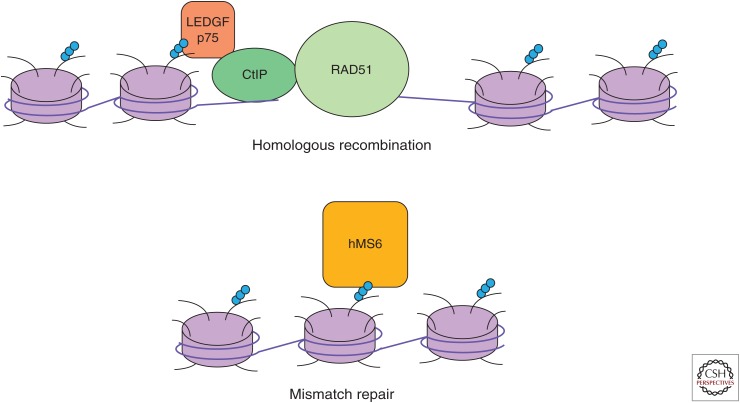
Substantial evidence supports the involvement of SETD2 and H3K36 methylation in DNA damage repair by homologous recombination (HR) (Fig. 4). SETD2 silencing resulted in a loss of HR at experimentally induced sites of double-strand breaks (DSBs) (Aymard et al. 2014). In particular, SETD2 loss decreased levels of ATM phosphorylation and consequently p53 phosphorylation (Carvalho et al. 2014) with decreased levels of p53 transcriptional targets (Xie et al. 2008; Carvalho et al. 2014). SETD2 has been shown to associate with and potentially stabilize p53, which may partially account for the difference in transcriptional targets (Xie et al. 2008). SETD2 loss was also associated with decreased recruitment of the HR proteins 53BP1, RPA, and RAD51 to chromatin (Aymard et al. 2014; Pfister et al. 2014; Kanu et al. 2015). This effect seems to be mediated by the histone methylation activity of SETD2 since reintroduction of a catalytically dead SETD2 mutant failed to rescue RPA or RAD51 foci formation, and depletion of H3K36me3 by overexpression of the KDM4 demethylase or H3.3K36M also delayed RAD51 foci formation (Pfister et al. 2014).
Figure 4.
DNA damage repair. Specific H3K36me3 readers direct either homologous recombination (top) or mismatch repair (bottom).
LEDGF may bridge H3K36 methylation and the DNA damage response mechanism. In contrast to the role of the p52 isoform in transcription, the LEDGF/p75 isoform recruits carboxy-terminal binding protein and interacting protein (CtIP) to sites of DNA damage (Daugaard et al. 2012). CtIP processes DNA ends to enable binding of RAD51 (reviewed in Symington 2010). Depletion of SETD2 results in decreased LEDGF bound to chromatin, reduced CtIP recruitment to DSB and levels of single-stranded DNA near the DSB, suggesting impaired resection (Pfister et al. 2014). This suggests a model in which regions marked by H3K36me3 by SETD2 are bound by LEDGF following DSB, leading to recruitment of CtIP and RAD51 and, ultimately, repair by HR. Whether SETD2 is recruited to sites of DNA damage that will go on to repair by homologous recombination, or conversely, that homologous recombination is more likely at regions already marked by H3K36me3 remains unclear. In contrast to H3K36me3, H3K36me2 is rapidly induced after irradiation (Fnu et al. 2011). Increased levels of early nonhomologous end joining (NHEJ) factors were detected by immunoprecipitation of H3K36me2 following radiation.
In addition to HR, SETD2 is involved in mismatch repair (MMR) (Fig. 4). hMSH6, a component of the hMutSα complex that recognizes mismatches in the genome, also contains a PWWP domain that mediates interaction with methylated H3K36 (Li et al. 2013). SETD2 silencing or overexpression of KDM4 decreased MSH6 foci formation associated with increased microsatellite instability (MSI) (Li et al. 2013; Awwad and Ayoub 2015). However, ccRCC tumors samples with biallelic SETD2 loss did not show classic findings of MSI, increased breakpoints or a substantially increased mutation load, compared with tumor cells with monoallelic loss (Kanu et al. 2015). DNA breaks identified in tumors with monoalleic SETD2 loss showed significantly lower levels of H3K36me3. These data are consistent with the model that H3K36me3 marked sites are protected from breakage. Taken together, these studies suggest that H3K36 methylation functions in DNA damage repair with methylation status biasing toward different repair pathways, with dimethylation favoring NHEJ and trimethylation favoring HR and MMR. Although SETD2 deficient kidney cancers are not characterized by increased mutational level, it remains possible that intratumoral heterogeneity limits our ability to detect this feature (Gerlinger et al. 2012).
DNA METHYLATION AND REPLICATION
Alterations in DNA methylation have been linked to SETD2 and H3K36me3 loss. The DNA methyltransferases DNMT3A and DNMT3B contain a PWWP domain enabling binding to methylated H3K36 (Dhayalan et al. 2010; Baubec et al. 2015). DNMT3B was enriched at H3K36me3 marked gene bodies, and SETD2 knockout reduced DNMT3B binding (Baubec et al. 2015). In this study, SETD2 loss was associated with decreased de novo DNA methylation, although a separate study did not observe this association (Hahn et al. 2011). Alterations in DNA methylation correlated with SETD2 loss have been shown in ccRCC. In the TCGA analysis, SETD2 mutation was associated with decreased DNA methylation at regions that are normally marked by H3K36me3 in kidney (The Cancer Genome Atlas 2013a). Increased chromatin accessibility was also associated with regions of DNA hypomethylation in SETD2 mutant tumors (Simon et al. 2014). SETD2 loss has also been associated with increased DNA methylation at intergenic regions (Tiedemann et al. 2016). Overall, these data suggest that H3K36me3 directs DNA methyltransferases to gene bodies but in the absence of this histone modification, methylation increases elsewhere.
H3K36me3 levels are cell-cycle regulated with a peak in early S phase then declining to low levels that persist during G2/M (Li et al. 2013). This pattern suggests that SETD2 is most active during DNA replication. In support of a role during replication, depletion of SETD2 in kidney cancer cells slowed replication fork progression and led to an accumulation of cells in S phase (Kanu et al. 2015). However, in isogenic SETD2 knockout cell lines, cell-cycle differences were not observed (Pfister et al. 2015).
SETD2 IN CANCER DEVELOPMENT AND THERAPEUTICS
How SETD2 loss results in cancer development remains unknown. However, several studies have linked H3K36 methylation to aberrant differentiation or proliferation. SETD2 loss disrupts murine embryonic stem-cell differentiation, possibly by altering intracellular signaling (Zhang et al. 2014). Expression of H3.3 mutants that inhibit H3K36 methylation in chondrocytes and mesenchymal progenitor cells disrupted differentiation (Fang et al. 2016; Lu et al. 2016). Mouse mesenchymal progenitor cells (MPCs) that stably express either wild-type or K36M mutant H3.3 formed tumors after subcutaneous injection in immunocompromised mice (Lu et al. 2016). In renal primary epithelial tubule cells, cells considered to be the progenitor for ccRCC, SETD2 knockdown resulted in continued proliferation well past the point at which these cells typically senesce (Li et al. 2016). In models of MLL-rearranged leukemia, SETD2 loss is associated with increased colony formation, proliferation, and accelerated leukemia development after transplantation (Zhu et al. 2014). Taken together, these studies support a role of SETD2 in facilitating faithful differentiation. Interestingly, germline mutations in SETD2 as well as NSD1 (the H3K36 dimethylase; see Fig. 1) have been associated with Sotos and Sotos-like overgrowth syndromes (Kurotaki et al. 2002; Luscan et al. 2014; Tlemsani et al. 2016). Sotos syndrome has been associated with an increased frequency of malignancy, particularly acute leukemias and lymphomas, Wilms tumor, and neuroblastoma (reviewed in Lapunzina and Cohen 2005).
The clinical implications of SETD2 loss in cancer have primarily focused on ccRCC. SETD2 mutation is associated with worse cancer-specific survival in the TCGA dataset (Hakimi et al. 2013b). Additionally, SETD2 mutation was a univariate predicator of time to recurrence, and was found at higher percentages in late stage tumors. Tumors with BAP1, SETD2, or KDM5C mutation were more likely to present with advanced stage (Hakimi et al. 2013a). In metastatic RCC, low SETD2 expression was associated with reduced overall and progression-free survival, and was an independent prognostic marker for these endpoints (Wang et al. 2016). SETD2 expression was lower in breast tumors, compared with matched normal tissue (Al Sarakbi et al. 2009; Newbold and Mokbel 2010), with expression inversely correlated with increasing tumor stage (Al Sarakbi et al. 2009). In these patients, SETD2 mRNA levels were lower in patients with poor outcomes, such as metastasis, local recurrence, and cancer-specific death.
Several studies have explored whether SETD2 loss sensitizes tumor cells to targeted agents. TGX221, a selective PI3Kβ inhibitor, was selectively toxic to RCC cells that were mutant for both VHL and SETD2, whereas cells lacking either mutation were not sensitive (Feng et al. 2015). Treatment with this compound resulted in decreased migration and invasion of mutant cell lines. Using a synthetic lethality screening strategy, H3K36me3-deficient cell lines were found to be sensitive to WEE1 inhibition (Pfister et al. 2015). The proposed target of this synthetic lethal interaction is RRM2, a ribonucleotide reductase subunit. SETD2 deficiency and WEE1 inhibition each decreased RRM2 levels, and the combination resulted in further depletion. WEE1 inhibition in the context of SETD2 deficiency critically reduces the dNTP pool causing cells to accumulate in nonreplicating S phase, replication stress, and cell death.
CONCLUDING REMARKS
Large sequencing studies have increasingly implicated mutations in epigenetic modifiers as critical events in cancer development and have identified SETD2 loss as a key feature of multiple types of cancer. SETD2 has been implicated in many chromatin-directed nuclear processes, including transcriptional regulation, DNA damage repair, DNA methylation, and replication. These effects are likely mediated by H3K36me3-binding reader proteins. Consequently, shifts in H3K36 di- and trimethylation are expected to lead to loss of appropriate reader targeting or redistribution. The relative importance of the H3K36-associated functions to cancer development remains unclear. Although the focus of SETD2 research has been primarily on histone regulation, it is possible that SETD2 may have important nonhistone targets.
As SETD2 mutation is associated with more aggressive cancer, it is important to fully understand the effect of SETD2 loss on oncogenesis. Vulnerabilities created by SETD2 through deregulated transcription and DNA replication may offer therapeutic strategies.
ACKNOWLEDGMENTS
We thank Brian D. Strahl for critical reading and feedback. This project is supported by University of North Carolina (UNC) Medical Scientist Training Program T32 GM008719-12 and National Research Service Award (NRSA) award F30 CA192643-02 (C.C.F.) and R01 CA198482 and R01 CA166447 (I.J.D.).
Footnotes
Editors: Scott A. Armstrong, Steven Henikoff, and Christopher R. Vakoc
Additional Perspectives on Chromatin Deregulation in Cancer available at www.perspectivesinmedicine.org
REFERENCES
- Al Sarakbi W, Sasi W, Jiang WG, Roberts T, Newbold RF, Mokbel K. 2009. The mRNA expression of SETD2 in human breast cancer: Correlation with clinico-pathological parameters. BMC Cancer 9: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awwad SW, Ayoub N. 2015. Overexpression of KDM4 lysine demethylases disrupts the integrity of the DNA mismatch repair pathway. Biol Open 4: 498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iacovoni JS, Daburon V, Miller KM, Jackson SP, et al. 2014. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 21: 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers F a, Thorne AW, Crane-Robinson C, Kouzarides T. 2005. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 280: 17732–17736. [DOI] [PubMed] [Google Scholar]
- Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, Schübeler D. 2015. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520: 243–247. [DOI] [PubMed] [Google Scholar]
- Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, Wedge DC, Cooke SL, Gundem G, Davies H, et al. 2013. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet 45: 1479–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. 2012. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485: 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia W-J, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592. [DOI] [PubMed] [Google Scholar]
- Carvalho S, Raposo AC, Martins FB, Grosso AR, Sridhara SC, Rino J, Carmo-Fonseca M, de Almeida SF. 2013. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res 41: 2881–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S, Vítor AACA, Sridhara SCS, Martins FB, Raposo AC, Desterro JMP, Ferreira J, de Almeida SF, Filipa BM, Ana CR, et al. 2014. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. eLife 3: e02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian Y, Hillringhaus L, Bagg EA, Rose NR, Leung IKH, Li XS, Woon ECY, Yang M, et al. 2011. Scientific report. Nat Publ Gr 12: 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. 2010. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugaard M, Baude A, Fugger K, Povlsen LK, Beck H, Sørensen CS, Petersen NHT, Sorensen PHB, Lukas C, Bartek J, et al. 2012. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat Struct Mol Biol 19: 803–810. [DOI] [PubMed] [Google Scholar]
- de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. 2011. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol 18: 977–983. [DOI] [PubMed] [Google Scholar]
- Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. 2010. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem 285: 26114–26120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duns G, Hofstra RMW, Sietzema JG, Hollema H, van Duivenbode I, Kuik A, Giezen C, Jan O, Bergsma JJ, Bijnen H, et al. 2012. Targeted exome sequencing in clear cell renal cell carcinoma tumors suggests aberrant chromatin regulation as a crucial step in ccRCC development. Hum Mutat 33: 1059–1062. [DOI] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. 2008. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J 27: 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Gan H, Lee J, Han J, Wang Z, Riester SM, Jin L, Chen J, Zhou H, Wang J, et al. 2016. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352: 1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Ding G, Jiang H, Ding Q, Wen H. 2015. Loss of MLH1 confers resistance to PI3Kβ inhibitors in renal clear cell carcinoma with SETD2 mutation. Tumour Biol 36: 3457–3464. [DOI] [PubMed] [Google Scholar]
- Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. 2011. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci 108: 540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, Liu XY, Sturm D, Korshunov A, Jones DTW, Witt H, Kool M, Albrecht S, et al. 2013. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol 125: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YG, Yang H, Zhao J, Jiang YJ, Hu HY. 2014. Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin. Structure 22: 378–386. [DOI] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossage L, Murtaza M, Slatter AF, Lichtenstein CP, Warren A, Haynes B, Marass F, Roberts I, Shanahan SJ, Claas A, et al. 2014. Clinical and pathological impact of VHL, PBRM1, BAP1, SETD2, KDM6A, and JARID1c in clear cell renal cell carcinoma. Genes Chromosomes Cancer 53: 38–51. [DOI] [PubMed] [Google Scholar]
- Grosso AR, Leite AP, Carvalho S, Matos MR, Martins FB, Vítor AC, Desterro JMP, Carmo-Fonseca M, de Almeida SF. 2015. Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. eLife 4: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Zheng L, Park JW, Lv R, Chen H, Jiao F, Xu W, Mu S, Wen H, Qiu J, et al. 2014. BS69/ZMYND11 reads and connects histone H3.3 Lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol Cell 56: 298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Wu X, Li AX, Hahn T, Pfeifer GP. 2011. Relationship between gene body DNA methylation and intragenic H3K9me3 and H3K36me3 chromatin marks. PLoS ONE 6: e18844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi AA, Chen Y-B, Wren J, Gonen M, Abdel-Wahab O, Heguy A, Liu H, Takeda S, Tickoo SK, Reuter VE, et al. 2013a. Clinical and pathologic impact of select chromatin-modulating tumor suppressors in clear cell renal cell carcinoma. Eur Urol 63: 848–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi AA, Ostrovnaya I, Reva B, Schultz N, Chen Y-B, Gonen M, Liu H, Takeda S, Voss MH, Tickoo SK, et al. 2013b. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: A report by MSKCC and the KIRC TCGA research network. Clin Cancer Res 19: 3259–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Park IY, Zhao H, Tong P, Champion MD, Yan H, Monzon F, Hoang, Tamboli P, Parker S, et al. 2015. High-resolution profiling of histone h3 lysine 36 trimethylation in metastatic renal cell carcinoma. Oncogene 35: 1565–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov G V, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. 2012. A landscape of driver mutations in melanoma. Cell 150: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KK, McPherson JR, Tay ST, Das K, Tan IB, Ng CCY, Chia N-Y, Zhang SL, Myint SS, Hu L, et al. 2015. SETD2 histone modifier loss in aggressive GI stromal tumours. Gut doi: 10.1136/gutjnl-2015-309482. [DOI] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. 2012. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150: 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Stransky N, McCord CL, Cerami E, Lagowski J, Kelly D, Angiuoli S V, Sausen M, Kann L, Shukla M, et al. 2014. Genomic analyses of gynaecologic carcinosarcomas reveal frequent mutations in chromatin remodelling genes. Nat Commun 5: 5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Lee D, Lee J, Park D, Kim YJ, Park WY, Hong D, Park PJ, Lee E. 2015. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet 47: 1242–1248. [DOI] [PubMed] [Google Scholar]
- Kanu N, Grönroos E, Martinez P, Burrell Ra, Yi Goh X, Bartkova J, Maya-Mendoza A, Mistrík M, Rowan J, Patel H, et al. 2015. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene 34: 5699–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim H, Fong N, Erickson B, Bentley DL. 2011. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci 108: 13564–13569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. 2009. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet 41: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 23: 4207–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotaki N, Imaizumi K, Harada N, Masuno M, Kondoh T, Nagai T, Ohashi H, Naritomi K, Tsukahara M, Makita Y, et al. 2002. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet 30: 365–366. [DOI] [PubMed] [Google Scholar]
- Lapunzina P, Cohen MM. 2005. Risk of tumorigenesis in overgrowth syndromes: A comprehensive review. Am J Med Genet-Semin Med Genet 137C: 53–71. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Sholl LM, Lindeman NI, Granter SR, Laga AC, Shivdasani P, Chin G, Luke JJ, Ott PA, Hodi FS, et al. 2015. Targeted next-generation sequencing reveals high frequency of mutations in epigenetic regulators across treatment-naïve patient melanomas. Clin Epigenetics 7: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski L a, Garcia B a, Muir TW, Becher OJ, Allis CD. 2013. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR, Workman JL. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 278: 8897–8903. [DOI] [PubMed] [Google Scholar]
- Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P. 2005. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc Natl Acad Sci 102: 17636–17641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. 2007. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 21: 1422–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li G-M. 2013. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell 153: 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Hanna GJ, Laga AC, Haddad RI, Lorch JH, Hammerman PS. 2015. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin Cancer Res 21: 1447–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kluiver J, Osinga J, Westers H, van Werkhoven MB, Seelen MA, Sijmons RH, van den Berg A, Kok K. 2016. Functional studies on primary tubular epithelial cells indicate a tumor suppressor role of SETD2 in clear cell renal cell carcinoma. Neoplasia 18: 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickwar CR, Rao B, Shabalin Aa, Nobel AB, Strahl BD, Lieb JD. 2009. The set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS ONE 4: e4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S-Y, Joseph NM, Ravindranathan A, Stohr BA, Greenland NY, Vohra P, Hosfield E, Yeh I, Talevich E, Onodera C, et al. 2016. Genomic profiling of malignant phyllodes tumors reveals aberrations in FGFR1 and PI-3 kinase/RAS signaling pathways and provides insights into intratumoral heterogeneity. Mod Pathol 29: 1012–1027. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Zhou XZ, Shen M, Lu KP. 1999. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283: 1325–1328. [DOI] [PubMed] [Google Scholar]
- Lu C, Ward P, Kapoor G, Rohle D. 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Jain SU, Hoelper D, Bechet D, Molden RC, Ran L, Murphy D, Venneti S, Hameed M, Pawel BR, et al. 2016. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352: 844–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. 2010. Regulation of alternative splicing by histone modifications. Science 327: 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscan A, Laurendeau I, Malan V, Francannet C, Odent S, Giuliano F, Lacombe D, Touraine R, Vidaud M, Pasmant E, et al. 2014. Mutations in SETD2 cause a novel overgrowth condition. J Med Genet 51: 512–517. [DOI] [PubMed] [Google Scholar]
- Mar BG, Bullinger LB, McLean KM, Grauman P V, Harris MH, Stevenson K, Neuberg DS, Sinha AU, Sallan SE, Silverman LB, et al. 2014. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun 5: 3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Kim J, Wang GG, Stadler SC, Basrur V, Whitcomb SJ, Wang Z, Ruthenburg AJ, Elenitoba-Johnson KSJ, Roeder RG, et al. 2010. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell 38: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold RF, Mokbel K. 2010. Evidence for a tumour suppressor function of SETD2 in human breast cancer: A new hypothesis. Anticancer Res 30: 3309–3311. [PubMed] [Google Scholar]
- Parker H, Rose-Zerilli MJJ, Larrayoz M, Clifford R, Edelmann J, Blakemore S, Gibson J, Wang J, Ljungström V, Wojdacz TK, et al. 2016. Genomic disruption of the histone methyltransferase SETD2 in chronic lymphocytic leukaemia. Leukemia doi: 10.1038/leu.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ, Helleday T, Legube G, LaThangue NB, Porter ACG, et al. 2014. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep 7: 2006–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister SX, Markkanen E, Jiang Y, Sarkar S, Woodcock M, Orlando G, Mavrommati I, Pai C-C, Zalmas L-P, Drobnitzky N, et al. 2015. Inhibiting WEE1 selectively kills histone H3K36me3-deficient cancers by dNTP starvation. Cancer Cell 28: 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore W a. 2012. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet 8: e1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G. 2009. Chromatin organization marks exon–intron structure. Nat Struct Mol Biol 16: 990–995. [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DTW, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DAK, Tönjes M, et al. 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231. [DOI] [PubMed] [Google Scholar]
- Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman V, Jaiswal BS, et al. 2012. Recurrent R-spondin fusions in colon cancer. Nature 488: 660–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Garrido M, Botton T, Talevich E, Yeh I, Sanborn JZ, Chung J, Wang NJ, Kakavand H, Mann GJ, et al. 2015. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat Genet 47: 1194–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JM, Hacker KE, Singh D, Brannon a R, Parker JS, Weiser M, Ho TH, Kuan PF, Jonasch E, Furey TS, et al. 2014. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res 24: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DP, Kimura A, Chylack LT, Shinohara T. 2000. Lens epithelium-derived growth factor (LEDGF/p75) and p52 are derived from a single gene by alternative splicing. Gene 242: 265–273. [DOI] [PubMed] [Google Scholar]
- Stolle C, Glenn G, Zbar B, Humphrey JS, Choyke P, Walther M, Pack S, Hurley K, Andrey C, Klausner R, et al. 1998. Improved detection of germline mutations in the von Hippel–Lindau disease tumor suppressor gene. Hum Mutat 12: 417–23. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22: 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DTW, Konermann C, Pfaff E, Tönjes M, Sill M, Bender S, et al. 2012. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22: 425–437. [DOI] [PubMed] [Google Scholar]
- Sun XJ, Wei J, Wu XY, Hu M, Wang L, Wang HH, Zhang QH, Chen SJ, Huang QH, Chen Z. 2005. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem 280: 35261–35271. [DOI] [PubMed] [Google Scholar]
- Symington LS. 2010. Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol 51: 195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Ong CK, Lim WK, Ng CCY, Thike AA, Ng LM, Rajasegaran V, Myint SS, Nagarajan S, Thangaraju S, et al. 2015. Genomic landscapes of breast fibroepithelial tumors. Nat Genet 47: 1341–1345. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas. 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas. 2013a. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas. 2013b. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas. 2014a. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513: 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas. 2014b. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas. 2014c. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas; Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, et al. 2013. Integrated genomic characterization of endometrial carcinoma. Nature 497: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas; Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Davis C, Wheeler DA, Murray BA, Schmidt L, et al. 2016. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med 374: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedemann RL, Hlady RA, Hanavan PD, Lake DF, Tibes R, Lee J-H, Choi J, Ho TH, Robertson KD. 2016. Dynamic reprogramming of DNA methylation in SETD2-deregulated renal cell carcinoma. Oncotarget 7: 1927–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlemsani C, Luscan A, Leulliot N, Bieth E, Afenjar A, Baujat G, Doco-Fenzy M, Goldenberg A, Lacombe D, Lambert L, et al. 2016. SETD2 and DNMT3A screen in the Sotos-like syndrome French cohort. J Med Genet doi: 10.1136/jmedgenet-2015-103638. [DOI] [PubMed] [Google Scholar]
- Van Allen EM, Mouw KW, Kim P, Iyer G, Wagle N, Al-Ahmadie H, Zhu C, Ostrovnaya I, Kryukov GV, O'Connor KW, et al. 2014. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov 4: 1140–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. 2012. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489: 452–455. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. 2012. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu L, Qu Y, Xi W, Xia Y, Bai Q, Xiong Y, Long Q, Xu J, Guo J. 2016. Prognostic value of SETD2 expression in patients with metastatic renal cell carcinoma treated with tyrosine kinase inhibitors. J Urol 195: 1363–1370. [DOI] [PubMed] [Google Scholar]
- Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, et al. 2015. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 6: 6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. 2012. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44: 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, Zhu X, Qu C, Chen X, Zhang JJ, et al. 2014. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 46: 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang W, He J, Huang C, Chen L, Tao D, Wu X. 2015. miR-106b-5p targets tumor suppressor gene SETD2 to inactive its function in clear cell renal cell carcinoma. Oncotarget 6: 4066–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev 17: 654–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P, Tian C, An L, Nie J, Lu K, Xing G, Zhang L, He F. 2008. Histone methyltransferase protein SETD2 interacts with p53 and selectively regulates its downstream genes. Cell Signal 20: 1671–1678. [DOI] [PubMed] [Google Scholar]
- Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S. 2011. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J 30: 1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S, Ito S, Yang C, Wang P. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Lucas JS, Jones KA. 2008. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev 22: 3422–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. 2010. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 17: 198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xie J, Long C, Erdjument-Bromage H, Ding X, Zheng Y, Tempst P, Chen S, Zhu B, Reinberg D. 2009. Heterogeneous nuclear ribonucleoprotein L is a subunit of human KMT3a/set2 complex required for H3 Lys-36 trimethylation activity in vivo. J Biol Chem 284: 15701–15707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbar B, Brauch H, Talmadge C, Linehan M. 1987. Loss of alleles of loci on the short arm of chromosome 3 in renal cell carcinoma. Nature 327: 721–724. [DOI] [PubMed] [Google Scholar]
- Zhang P, Du J, Sun B, Dong X, Xu G, Zhou J, Huang Q, Liu Q, Hao Q, Ding J. 2006. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res 34: 6621–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xie S, Zhou Y, Xie Y, Liu P, Sun M, Xiao H, Jin Y, Sun X, Chen Z, et al. 2014. H3K36 histone methyltransferase Setd2 is required for murine embryonic stem cell differentiation toward endoderm. Cell Rep 8: 1989–2002. [DOI] [PubMed] [Google Scholar]
- Zhu X, He F, Zeng H, Ling S, Chen A, Wang Y, Yan X, Wei W, Pang Y, Cheng H, et al. 2014. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet 46: 287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]