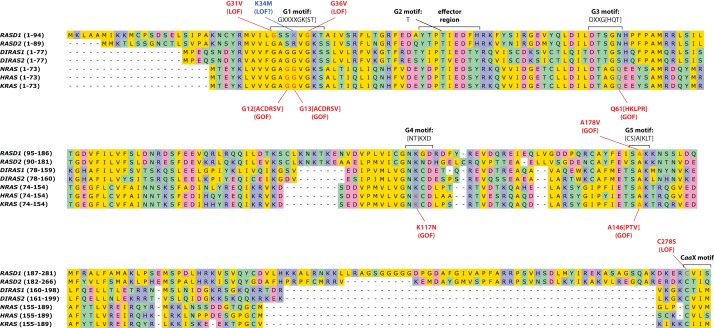
Figure 4.
Multiple sequence alignment of select proteins in the Ras family of small monomeric GTPases to which RASD1 belongs (Wennerberg et al. 2005). Indicated motifs (taken from Wennerberg et al. 2005 and Bourne et al. 1991), and then verified via UniProt (The UniProt Consortium et al. 2015; retrieved 2016-09-24): GDP/GTP-binding G-box motifs (G1–G5); effector region; CaaX amino-terminal motif that undergoes posttranslational modification (a denotes any aliphatic amino acid). RASD2, encoding the protein Rhes, is shown because it is the closest human homolog to RASD1 (63% protein sequence identity); the two form the RASD subfamily that is distinct from other Ras family proteins (<45% protein sequence identity). DIRAS1 and DIRAS2 are shown because they are the next closest homologs to RASD family proteins. NRAS, HRAS, and KRAS are shown because they are well-characterized oncogenes. Functional impact of the RASD1 mutations (in red) has been demonstrated experimentally: p.G31V (Cismowski et al. 1999, 2000; Vaidyanathan et al. 2004), p.G36V (Cismowski et al. 1999), p.A178V (Graham et al. 2001), and p.C278S (Graham et al. 2001; Vaidyanathan et al. 2004). Mutations in NRAS, HRAS, and KRAS (in red) are widely known oncogenic mutations and are also recurrent somatic mutations across multiple neoplasm types in COSMIC (Forbes et al. 2015) (accessed 2016-09-25), except for HRAS amino acid A146 (no mutations of any type in COSMIC, although p.A146V results in constitutive activation [Feig and Cooper 1988] and may be germline pathogenic in Costello syndrome, ClinVar accession RCV000013445.18), and NRAS amino acid K117 (no mutations of any type in COSMIC and no published evidence on any K117 mutation). Amino acid ranges are given in parentheses next to the gene symbols. Amino acids are color-coded according to biochemical class (yellow, nonpolar; green, polar; blue, basic; pink, acidic). For positions where one of several amino acids is possible, the possibilities are given in brackets. X, any amino acid; LOF, loss of function; GOF, gain of function.