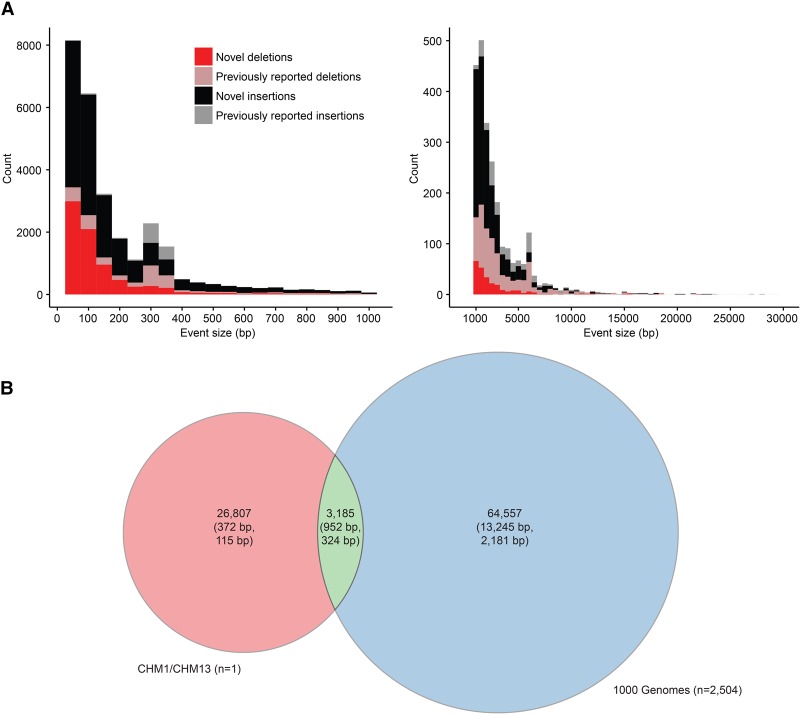
Figure 1.
Structural variant (SV) discovery. (A) SV deletions (red) and insertions (black) identified by SMRT-SV in a theoretical diploid human (CHM1 and CHM13) are classified as either novel (83%) or previously reported (17%) based on their presence in previously published SV call sets (Conrad et al. 2010; Kidd et al. 2010a; Mills et al. 2011; Sudmant et al. 2015a,b). (B) Compared specifically against insertions and deletions from Phase 3 of the 1000 Genomes Project (Sudmant et al. 2015b). Counts per call set are shown with mean and median SV size (base pair) shown in parentheses. The Venn diagram compares one theoretical diploid genome sequenced and analyzed using SMRT sequence data versus 2504 diploid genomes lightly sequenced (approximately sixfold coverage) with Illumina sequence.