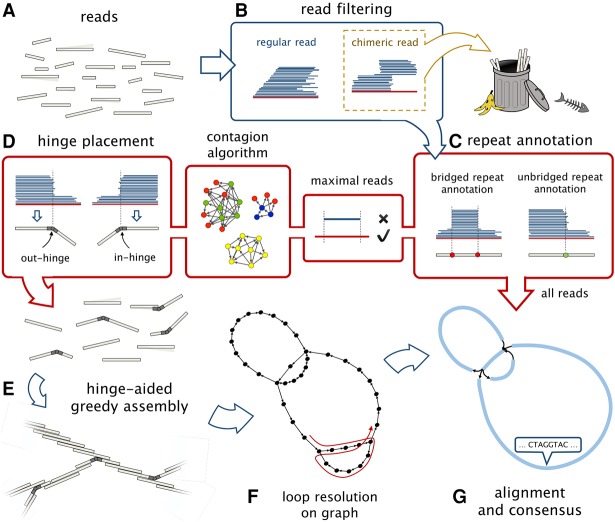
Figure 3.
HINGE pipeline. (A) The input to the HINGE pipeline is a set of long error-prone reads. (B) Chimeric reads are detected through their pile-o-grams and are discarded. (C) The beginning/end of repeats are annotated on the reads. This is done by detecting a sharp increase/decrease in the number of alignments on a read. The repeat annotations are also identified as bridged or unbridged. (D) Maximal reads (i.e., reads that are not contained in other reads) are selected and fed to the Contagion algorithm, which is responsible for spreading the information about which repeats are bridged to all the reads, allowing us to place exactly one in-hinge and one out-hinge on the reads that originated from unbridged occurrences of a repeat. (E) The set of maximal reads (some of which are not hinged) is the input to the hinge-aided greedy assembly. (F) After obtaining the read-overlap graph, we resolve repeats that admit only one traversal. (G) Finally, by mapping all the reads onto the resulting overlap graph, we use standard consensus methods to generate contigs.