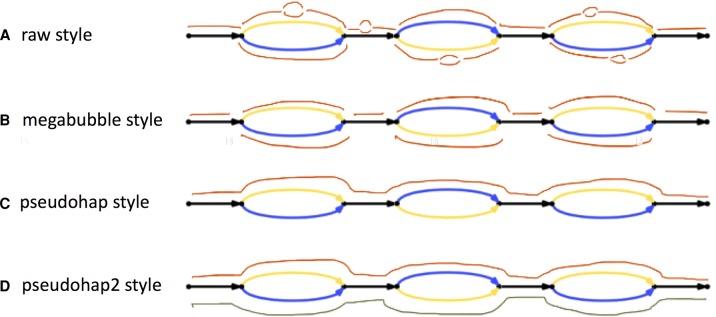
Figure 3.
Representation of Supernova assemblies as FASTA. Several styles are depicted. (A) The raw style represents every edge in the assembly as a FASTA record (red segments). These include microbubble arms and also gaps (printed as records comprising 100 Ns for gaps bridged by read pairs, or a larger number, the estimated gap size) (Supplemental Note 5). Unresolved cycles are replaced by a path through the cycle, followed by 10 Ns. Bubbles and gaps generally appear once per 10–20 kb; consequently, FASTA records from A are much shorter (∼100 times) than those from B, C, and D. For each edge in the raw graph, there is also an edge written to the FASTA file representing the reverse complement sequence. For the remaining output styles, we flatten each microbubble by selecting the branch having highest coverage, merge gaps with adjacent sequences (leaving Ns), and drop reverse complement edges. (B) In this style each megabubble arm corresponds to a FASTA record, as does each intervening sequence. (C) The pseudohap style generates a single record per scaffold. As compared to the megabubble style, in the example, seven red edges are seen on top (corresponding to seven FASTA records) that are combined into a single FASTA record in the pseudohap style. Megabubble arms are chosen arbitrarily so many records will mix maternal and paternal alleles. (D) This style is like the pseudohap option, except that for each scaffold, two “parallel” pseudohaplotypes are created and placed in separate FASTA files.