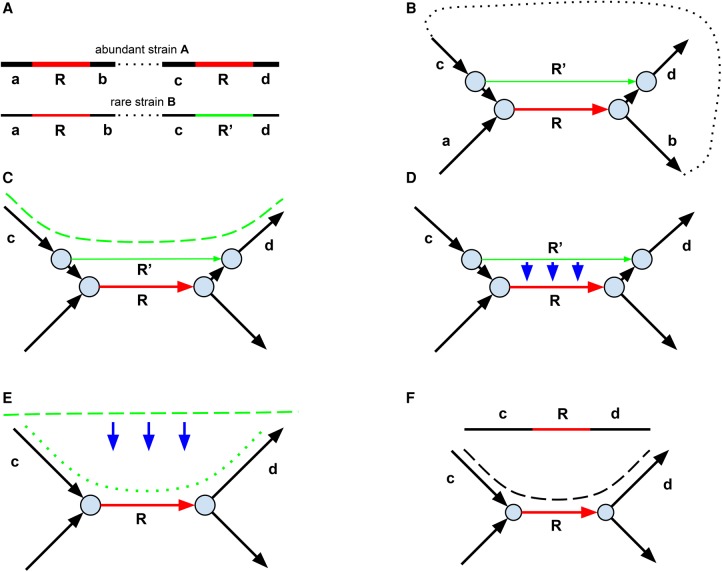
Figure 5.
Repeat resolution in metagenomic assembly. (A) One of two identical copies of a long (longer than the insert size) repeat R (red) in the abundant strain has mutated into a unique genomic “green” region R′ in the rare strain. (B) The assembly graph resulting from a mixture of reads from the abundant and rare strains. Two alternative paths between the start and the end of the green edge (one formed by a single green edge and another formed by two black and one red edge) form a bulge. (C) The strain-contig spanning R′ (shown by green dashed line) constructed by exSPAnder at the “generating strain-contigs” step. (D) Masking of the strain variation at the “transforming assembly graph into consensus assembly graph” step leads to a projection of a bulge (formed by red and green edges) and results in the consensus assembly graph shown in E. The blue arrows emphasize that SPAdes projects rather than deletes bulges, facilitating the subsequent reconstruction of strain-paths in the consensus assembly graph. (E) Reconstruction of the strain-path (green dotted line), corresponding to a strain-contig (green dashed line) at the “generating strain-paths in the consensus assembly graph” step. (F) At the “repeat resolution using strain-paths” step, metaSPAdes utilizes both strain-paths and paired reads to resolve repeats in the consensus graph. The green dotted strain-path from E is used as additional information to reconstruct the consensus contig cRd spanning the long repeat.