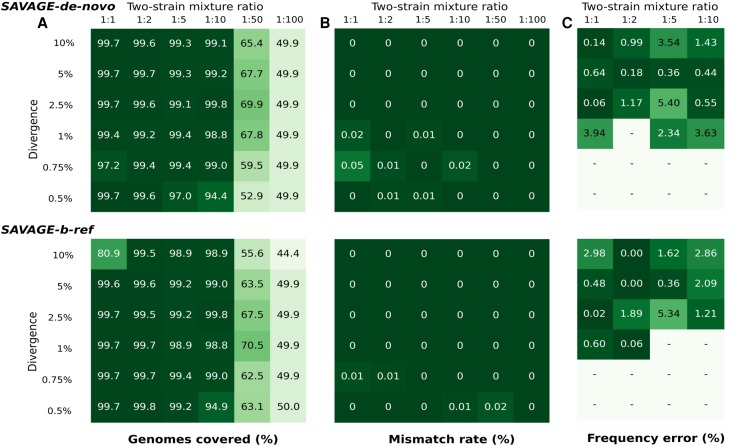
Figure 3.
Performance of SAVAGE-de-novo and SAVAGE-b-ref, depending on pairwise distance and mixture ratio. (A) Target genome fraction recovered (%) considering all maximally extended contigs ≥500 bp. (B) Overall mismatch rate (%) considering all maximally extended contigs ≥500 bp. (C) Relative error of estimated frequency for the minor strain (%). Frequency estimates were computed using Kallisto, and only assemblies containing exactly two maximally extended contigs longer than 4000 bp were evaluated.