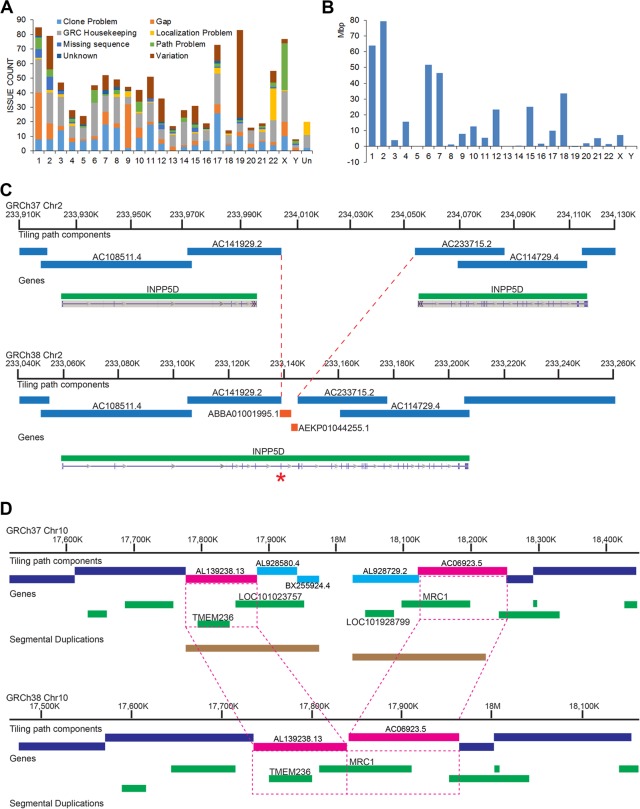
Figure 1.
Summary of GRCh38 updates. (A) Chart showing issues resolved for GRCh38 on each chromosome by issue type. Each issue represents a unique assembly evaluation and corresponding curation decision. (B) Changes in placed scaffold N50 length from GRCh37 to GRCh38. Changes on Chromosomes 5, 13, 19, and Y are <55 kbp each. (C) Addition of whole-genome sequencing components (orange bars) resolves a GRCh37 gap, consolidating the split annotation of INPP5D and restoring a missing exon (asterisk) in GRCh38. The default 50-kbp gap in GRCh37 greatly overestimates the actual amount of missing sequence (∼6 kbp). (D) Schematic of a curated collapse in GRCh38 Chr 10. Clones from two incompatible haplotypes (pink and light blue) were mixed in the GRCh37 tiling path, creating a false gap and segmental duplication involving the single copy genes TMEM236 and MRC1 (top). In GRCh38 (bottom), clones from the blue haplotype have been eliminated (∼200 kbp), closing the gap and providing the correct gene content.