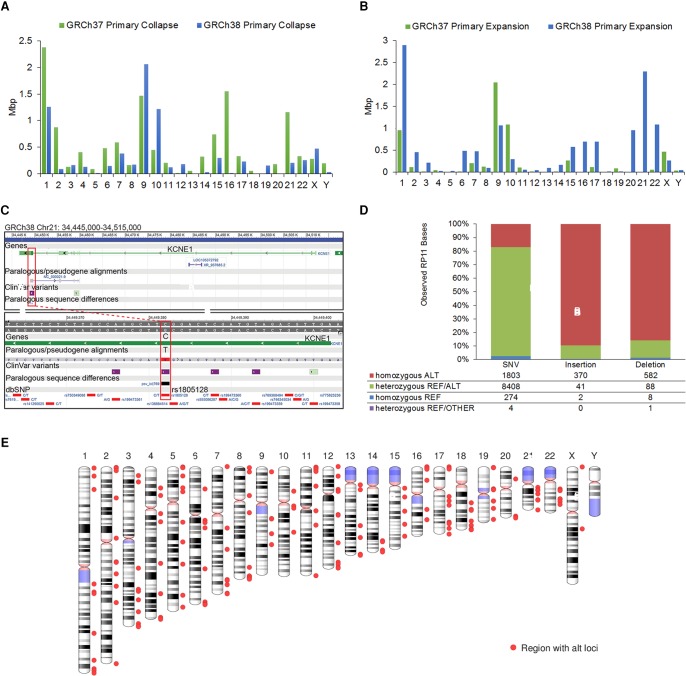
Figure 2.
Evaluation of assembly updates. (A,B) Plots showing the per-chromosome lengths of sequence collapse (A) and expansion (B) of the GRCh37 (green) and GRCh38 (blue) primary assembly units (from which alternate loci are excluded), based on their assembly–assembly alignment. (C) Browser view of KCNE1 on GRCh38 Chr 21. The lower panel shows a zoomed view of the top, illustrating a paralogous sequence alignment and paralogous variant (psv) overlapping SNP rs1805128 (red box), a putatively pathogenic ClinVar variant we observed remapping to multiple locations in GRCh38, due to the addition of paralogous sequence. Because previous assembly versions lack this paralog, reads may map incorrectly in this region, and the pathogenicity of the variant and associated diagnostic calls should not be based only on such analyses. (D) Plot showing the allele distribution in RP11 WGS reads for the set of GRCh37 bases located in RP11 assembly components that were flagged as putative errors because they were not observed in the 1000 Genomes phase 1 data set. (E) Ideogram showing the distribution of regions containing alternate loci scaffolds in GRCh38.