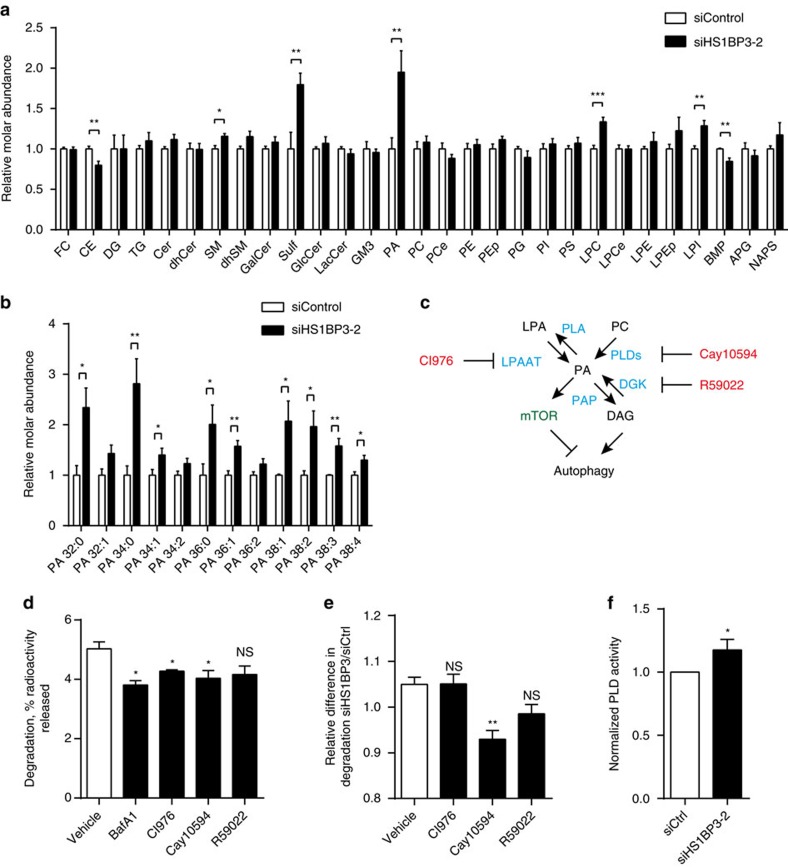
Figure 5. Autophagy is dependent on PA synthesis and HS1BP3 affects PLD activity.
(a) HEK cells were treated with non-targeting or HS1BP3 siRNA and starved for 2 h. The total lipid content was extracted and analysed by mass spectrometry. Only lipid species that were present in all experimental runs were included. The measured lipid concentrations were first normalized to total lipid per sample to determine molar percentages for each lipid subclass and species, before normalizing to the average molar percentage of controls (mean±s.e.m., n=6). For a full list of all lipids with abbreviations, see Supplementary Table 1. The lipidomics data sets have been deposited in the Dryad Digital Repository (doi:10.5061/dryad.gq3fk). (b) The relative molar abundance of all 12 PA species is shown. (mean±s.e.m., n=6). (c) Schematic overview of the enzymes (blue) governing the generation and turnover of PA from other lipid species (black) and the drugs (red) used to inhibit these pathways. (d) Degradation of long-lived proteins in HEK cells was quantified as the release of 14C-valine after 4 h starvation in the presence of the indicated inhibitors (mean±s.e.m., n=3). (e) The relative difference in long-lived protein degradation between HEK cells with siCtrl and siHS1BP3 was measured for the indicated inhibitors (mean±s.e.m., n=3). (f) PLD activity was measured in HEK lysates of cells transfected with non-targeting or HS1BP3 siRNA (mean±s.e.m., n=3). *P<0.05, **P<0.01, by Student's t-test.