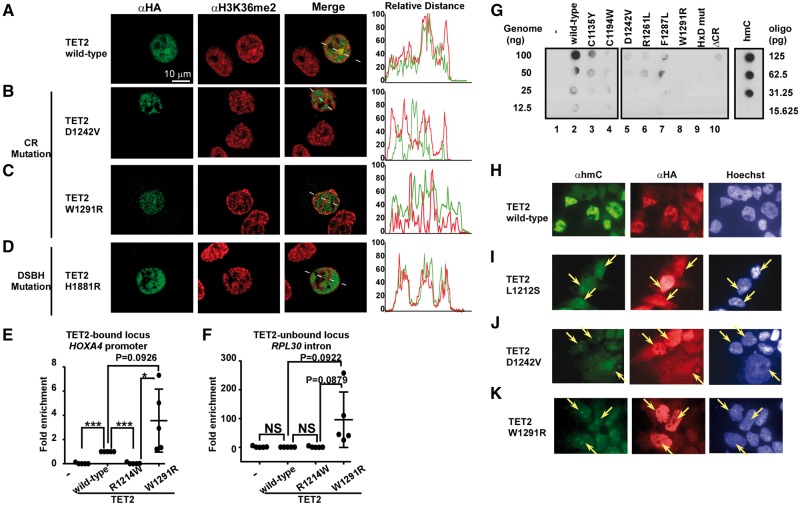
Fig. 2.
TET2 CR domain mutations disrupt the recognition of H3K36 methylation, its cellular localization, and enzyme activity in vivo. (A–D) Wild-type TET2 (A) and the DSBH domain mutant (H1881R) (D) but not CR domain mutants (B and C) localize to H3K36me2 loci. HeLa cells were transfected with the indicated expression plasmids and analysed by immunostaining using confocal fluorescence microscopy. Line profile analysis of colocalization between TET2 proteins and H3K36me2 is shown in the panel on the right. The white bar indicates 10 μm. (E and F) CR mutations disrupt the correct localization of TET2-bound (E) and TET2-unbound (F) loci. HEK293T cells were transfected with the indicated expression plasmids and analysed by ChIP-qPCR. *P<0.05; ***P<0.0001 (student t-test). (G) TET2 mutations in the CR domain alter enzymatic activity in cells. Dot-blot analysis of 5-hmC levels in HEK293T cells over-expressing wild-type or mutant TET2. Oligonucleotides containing hmC or DNA from HEK293T cells transfected with TET2 containing mutations in the HxD motif (TET2-HxDmut) were used for positive and negative controls, respectively. (H–K). Immunocytochemical detection of 5-hmC (green) and HA-TET2 (red) in HEK293T cells transiently transfected with wild-type or mutant TET2. Arrows indicate signature nuclear dot patterns and euchromatin regions.