Abstract
BACKGROUND
Detecting tumor-derived cell-free DNA (cfDNA) in the blood of brain tumor patients is challenging, presumably owing to the blood–brain barrier. Cerebral spinal fluid (CSF) may serve as an alternative “liquid biopsy” of brain tumors by enabling measurement of circulating DNA within CSF to characterize tumor-specific mutations. Many aspects about the characteristics and detectability of tumor mutations in CSF remain undetermined.
METHODS
We used digital PCR and targeted amplicon sequencing to quantify tumor mutations in the cfDNA of CSF and plasma collected from 7 patients with solid brain tumors. Also, we applied cancer panel sequencing to globally characterize the somatic mutation profile from the CSF of 1 patient with suspected leptomeningeal disease.
RESULTS
We detected tumor mutations in CSF samples from 6 of 7 patients with solid brain tumors. The concentration of the tumor mutant alleles varied widely between patients, from <5 to nearly 3000 copies/mL CSF. We identified 7 somatic mutations from the CSF of a patient with leptomeningeal disease by use of cancer panel sequencing, and the result was concordant with genetic testing on the primary tumor biopsy.
CONCLUSIONS
Tumor mutations were detectable in cfDNA from the CSF of patients with different primary and metastatic brain tumors. We designed 2 strategies to characterize tumor mutations in CSF for potential clinical diagnosis: the targeted detection of known driver mutations to monitor brain metastasis and the global characterization of genomic aberrations to direct personalized cancer care.
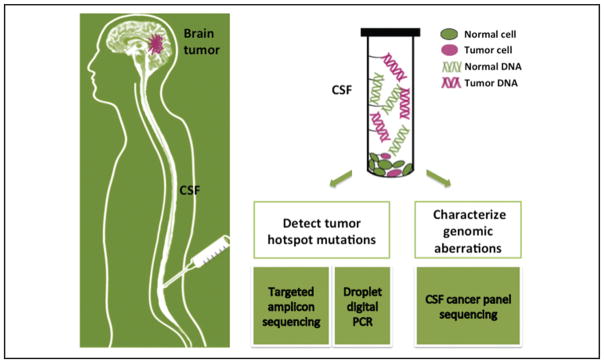
Recent studies have demonstrated great potential for using circulating cell-free DNA (cfDNA)5 in blood for cancer diagnosis, prognosis, and directed treatment (1). Although tumor-derived circulating cfDNA has been detected in a variety of cancers, it is rarely found in patients with isolated brain tumors, presumably owing to the blood–brain barrier (2). Determining the mutation profile of a brain tumor currently requires a risky and invasive intracranial biopsy. Less invasive strategies to diagnose central nervous system (CNS) tumors include magnetic resonance imaging scans or cytologic examination of cerebral spinal fluid (CSF); both are limited by low sensitivity and specificity (3). Neither provides information about the genetic alterations of the tumor. Because CSF circulates through the CNS and has a large interface with the brain and malignant tissues, CSF has a clear potential to carry tumor cfDNA and circulating tumor cells. Although cytology requires morphologically intact tumor cells for positive findings, cfDNA can presumably originate from dying but not circulating tumor cells anatomically distant from the site of CSF collection. Sampling CSF could therefore serve as a “liquid biopsy” to characterize the genomic aberrations of certain primary and metastatic brain tumors. A few studies have examined the nucleic acids in the CSF of brain tumor patients by use of PCR-based methods (4–7), but the characteristics of CSF tumor cfDNA have not been comprehensively investigated by the more sensitive and informative approach of high-throughput sequencing. Here, we designed 2 strategies to detect tumor mutations in CSF (Fig. 1). The first strategy quantifies tumor-specific hotspot mutations (the frequently mutated loci for a particular cancer type) in CSF by use of droplet digital PCR (ddPCR) or targeted amplicon sequencing. The second strategy comprehensively characterizes genomic aberrations in known cancer genes by CSF cancer panel sequencing, which is a method that enriches and sequences DNA from known cancer genes. The objectives of this study were to determine the presence and quantity of tumor cfDNA in CSF from patients with different types of brain tumors and to develop a minimally invasive method for diagnosing, characterizing, and tracking CNS tumors by analyzing their DNA in CSF.
Fig. 1. Brain tumor genome analysis and diagnosis from CSF DNA.
Two strategies are used for CSF DNA analysis: (1) The cancer hotspot mutations are detected with target amplicon sequencing or droplet digital PCR. (2) The cancer genomic aberrations are globally characterized by CSF cancer panel sequencing.
Materials and Methods
SAMPLE COLLECTION
We collected and banked samples from patients at Stanford hospital after obtaining informed consent under a Stanford institutional review board–approved protocol. We selected 10 patient samples with different types of brain tumors (see Supplemental Table 1, which accompanies the online version of this article at http://www.clinchem.org/content/vol61/issue3). Seven patients (S1–S7) had solid brain tumors, and 3 patients (L1–L3) had leptomeningeal disease. For patients with solid brain tumors, the CSF, blood, and brain tumor tissue were collected at the time of surgical resection. CSF from patients with leptomeningeal disease was collected at the time of a clinically indicated lumbar puncture.
SAMPLE PROCESSING
Blood and CSF samples were processed within 2 h of collection. The blood samples were separated into plasma and blood cells by centrifugation at 1000g for 10 min at 4 °C. The plasma was then centrifuged at 10,000g for 10 min at 4 °C to remove residual cells. We processed CSF samples by different methods according to their subsequent analysis (see online Supplemental Fig. 1). CSF samples from patients with solid brain tumors (S1–S7) were centrifuged at 1000g for 10 min at 4 °C to collect the cfDNA in the supernatant for the subsequent ddPCR and target amplicon sequencing analysis. For 1 leptomeningeal disease patient (L1), the CSF sample was frozen directly without centrifugation for the subsequent cancer panel sequencing analysis. For patients L2 and L3, CSF samples were split into 2 aliquots with equal volume. One aliquot was frozen directly without centrifugation to collect total DNA (including both cfDNA and cellular DNA); the other aliquot was centrifuged at 1000g for 10 min at 4 °C to collect cfDNA. We used these centrifuged and noncentrifuged samples from L2 and L3 to compare the size distribution and concentration between cfDNA and total DNA in CSF. All processed samples were stored at −80 °C before DNA isolation.
DNA ISOLATION
We isolated CSF cfDNA or total DNA from 1–10 mL CSF with the QIAamp Circulating Nucleic Acid Kit (Qiagen), and we isolated plasma cfDNA from 1–5 mL plasma with the QIAamp Circulating Nucleic Acid Kit. The blood cells separated from the blood samples were used for isolation of healthy DNA with the QIAamp DNA Mini kit (Qiagen). We isolated DNA from matched tumor tissue with the DNeasy Blood & Tissue kit (Qiagen). The size distribution of extracted DNA was measured by the Fragment Analyzer with the DNF-493 High Sensitivity Large Fragment Analysis Kit (Advanced Analytical).
EXOME SEQUENCING FOR TUMOR AND HEALTHY DNA
We performed exome sequencing on DNA isolated from tumor and healthy tissue to identify hotspot tumor mutations before subsequent ddPCR and targeted amplicon sequencing. We used approximately 1 μg DNA from matched tumor and healthy tissue to prepare indexed Illumina libraries. The DNA was sheared to approximately 200 bp by sonication (S220 focused-ultrasonicator, Covaris) before library construction. The Illumina libraries were constructed with NEBNext DNA Library Prep Kit for Illumina (NEB). NimbleGen SeqCap EZ Human Exome Library (Roche) was used to capture DNA from exonic regions. Approximately 100 million reads per library were sequenced using 100-bp paired-end runs on HiSeq 2000 or NextSeq sequencing system (Illumina). We trimmed the raw FASTQ reads with Trimmomatic (8), and the trimmed reads were aligned to human reference genome (hg19 assembly) with BWA software (9). To minimize bias, we removed PCR duplicates with the Picard MarkDuplicates program. After deduplication, reads were piped through GATK (10) Indel Realignment and BaseRecalibration. The somatic single-nucleotide variants (SNVs) were identified with MuTect (11) and annotated with ANNOVAR software (12). Only nonsynonymous mutations in the exonic regions of known cancer genes (COSMIC, catalog of somatic mutations in cancer (13) cancer census gene list, http://cancer.sanger.ac.uk/cancergenome/projects/census/) were retained. The mutations that have been curated in the COSMIC database and validated as driver mutations in the literature were selected as hotspot mutations.
ddPCR
We performed ddPCR (14, 15) assays with QX100 Droplet Digital PCR System (Bio-Rad) according to the manufacturer’s instructions. We generated approximately 15000 droplets from each cfDNA sample. After PCR was performed on all droplets, a fluorescent droplet reader collected the FAM (mutant) and HEX (wild-type) signals of the probes in each droplet. The mutant allele concentration (CMUT) and wild-type allele concentration (CWT) in the final PCR mix were calculated with QuantaSoft software (Bio-Rad) in copies per microliter. The mutant allele fraction (AFdPCR) was calculated with the following equation:
| (1) |
The concentrations of mutant and wild-type alleles were calculated with the following equations:
| (2) |
| (3) |
where CMUT_ori is mutant allele concentration in original CSF or plasma (copies/mL); CWT_ori is wild-type allele concentration in original CSF or plasma (copies/mL); VO is the volume of CSF or plasma used to extract cfDNA (mL); VE is the volume of cfDNA elution generated from DNA extraction (μL); and VP is the volume of cfDNA used in final PCR mix. (The volume of final PCR mix is 20 μL.)
The cfDNA concentration (CcfDNA) in original CSF or plasma (ng/mL) was calculated with the following equation:
| (4) |
The mass of 1 haploid human genome is 0.003 ng.
The mean number of target DNA copies per droplet (λ) was calculated as follows:
| (5) |
| (6) |
where n is the number of droplets for 1 ddPCR reaction; k is the number of positive droplets counted for 1 ddPCR reaction; kMUT is the number of positive mutant droplets counted for 1 ddPCR reaction; and kWT is the number of positive mutant droplets counted for 1 ddPCR reaction. The λ value was evaluated over all CSF and plasma cfDNA samples and ranged from 0.002 to 0.259.
TARGETED AMPLICON SEQUENCING
We designed primer pairs with Primer3 (17) (see online Supplemental Table 3). The method for targeted amplicon sequencing was adapted from Safe-SeqS as previously described (18, 19). Safe-SeqS increases the sensitivity of target sequencing by assigning a unique identifier (UID) to each template molecule and redundant sequencing. Because the concentration level of CSF cfDNA was lower than the range of DNA template concentration for standard Safe-SeqS, a preamplification step was added to increase the copies of target DNA. In brief, this method included 3 rounds of PCR: 1) preamplification PCR to increase the copies of target DNA; 2) first-round PCR to assign a 14-base UID to each preamplified DNA molecule; and 3) second-round PCR to add Illumina sequencing adaptors and sample-specific index barcode sequences. All 3 rounds of PCR were amplified with TaqDNA Polymerase (Platinum TaqDNA Polymerase High Fidelity, Invitrogen) and performed on a standard thermal cycler (Tetrad, Bio-Rad). Preamplification reactions were carried out in a 50-μL PCR reaction containing 0.1 μmol/L of each forward and reverse target-specific primer and 1–40 μL CSF cfDNA or plasma cfDNA. Reactions were subjected to 15 cycles of amplification (94 °C 2 min and 15 cycles of 95 °C 30 s, 50 °C 2 min, 68 °C 1 min). The PCR products were purified with AMPure XP beads (Beckman Coulter) at a 1:1 ratio and eluted in 20 μL water. The purified preamplification products were added to PCR Master Mix and first-round PCR primers (0.5 μmol/L). Reactions were subjected to 2 cycles of amplification (94 °C 2 min and 2 cycles of 94 °C 30 s, 61 °C 2 min, 68 °C 1 min). The PCR products were purified with AMPure XP beads at a 1:1 ratio and eluted in 20 μL water. The purified first-PCR products were added to PCR Master Mix and second-round primers (0.5 μmol/L). Reactions were subjected to 20 cycles of amplification (94 °C 2 min and 20 cycles of 94 °C 30 s, 65 °C 30 s, 68 °C 1 min). The PCR products were purified with AMPure XP beads at a 1:0.8 ratio and eluted in 20 μL water. The purified second-round PCR products from different samples were pooled together and sequenced at 5000–50000 coverage on an Illumina MiSeq platform. Sequencing reads from different samples were demultiplexed by their index barcodes. The mutant allele fraction (AFseq) was calculated for each sample with the following equation:
| (7) |
where RMUT is the number of reads with mutant allele at that locus, and RWT is the number of reads with wild-type allele at that locus.
CSF CANCER PANEL SEQUENCING
Because a considerable number of tumor cells grow along the dura and meninges of patients with leptomeningeal disease, cellular DNA in CSF constitutes an important source of tumor DNA (20). We therefore used total DNA from the CSF of a patient with leptomeningeal disease (L1) for the cancer panel sequencing.
Matched CSF total DNA and healthy tissue DNA were fragmented to approximately 200 bp by sonication before library construction. The Illumina libraries were constructed with Hyper Library Preparation Kit (Kapa Biosystems). The targeted DNA was enriched by the NimbleGene SeqCap EZ Comprehensive Cancer Design kit (Roche). This kit targets 4 Mb of DNA coding regions within 578 cancer genes. These cancer genes were gathered from the Cancer Gene Census (Sanger) and NCBI Gene Tests databases. We then sequenced the enriched DNA library using 100-bp paired-end runs on an Illumina HiSeq 2000 or NextSeq. The somatic mutations in the CSF DNA were identified by the BWA-GATK-MuTect (9, 21) bioinformatics pipeline as described in “Exome Sequencing for Tumor and Healthy DNA” above.
Results
CHARACTERIZATION OF DNA IN CSF
Unlike blood, which has 3500–10500 white blood cells/μL, very few cells are present in CSF under routine conditions (0–5 cells/μL). The scarcity of cells in CSF may reduce the background noise from healthy DNA when detecting mutations. Inflammation or tumor metastasis can increase the number of cells present in CSF (22). There are 2 sources of DNA in CSF: the cfDNA and genomic DNA from cells. Fig. 2A shows an example of the size distribution of CSF total DNA (including both cfDNA and cellular DNA) from a patient with leptomeningeal disease (L3). The peaks at 160 and 340 bp match signatures of cfDNA from an apoptotic source, and the large-size DNA fragments match the usual size distribution of genomic DNA from white blood cells and possibly tumor cells in CSF. Whereas the total DNA was isolated directly from CSF, cfDNA was isolated after removing the cells by spinning down the CSF.
Fig. 2. The characteristics of CSF DNA.
(A), An example of the size distribution of CSF total DNA (from L3, a patient with leptomeningeal disease from breast cancer), as measured by Fragment Analyzer. The CSF total DNA was composed of cfDNA and cellular genomic DNA (gDNA). RFU, relative fluorescence unit. (B), cfDNA concentrations of CSF and plasma samples from patients S1–S7. The cfDNA concentration values are included in Table 1. The black dots in the plot indicate the values of individual samples. (C), The correlation of mutant allele fraction (AF) measured by ddPCR and targeted amplicon sequencing (AmpSeq). The dots in the plot represent the AF of S1_CSF [NF2 (neurofibromin 2 [merlin]) p.R57*], S1_plasma (NF2 p.R57*), S5_CSF (KRAS p.G12D and TP53 p.R282W), S5_plasma (KRAS p.G12D and TP53 p.R282W), S6_CSF (EGFR p.L585R), S7_CSF (EGFR p.E746_A750del), and S7_plasma (EGFR p.E746_A750del). The AF values are included in Table 1.
The concentration of cfDNA was quantified by digital PCR using the probes listed in online Supplemental Table 4. There was much less cfDNA than total DNA in CSF. For example, the concentrations of cfDNA and total DNA from the same CSF sample (L2) were 0.45 and 3 ng/mL, respectively. The median concentration of cfDNA in CSF (patients S1–S7) was 2.1 ng/mL, which was lower than the median concentration of 7.7 ng/mL in plasma (Fig. 2B). The concentration of CSF cfDNA spanned a wide range from 0.3 to 18.1 ng/mL, corresponding to 90–5400 haploid genome equivalents/mL sample.
DETECTION OF TUMOR MUTATIONS IN CSF cfDNA
Of the 7 patients with solid tumors, 2 (S6 and S7) had cancer-specific mutations identified with the SNaPshot assay (23) by the clinical pathology laboratory (see online Supplemental Table 1). For the other 5 patients (S1–S5), somatic mutations were identified by exome sequencing of matched tumor tissue and healthy lymphocyte cells. Across these 5 paired samples, 2834 SNVs were called by MuTect, 37 of which were nonsynonymous mutations in the exonic regions of known cancer genes (see online Supplemental Table 2). Among these 37 SNVs, 6 hotspot mutations were selected for subsequent analysis (see online Supplemental Table 1). We used 1 hotspot mutation per patient with commercially premade ddPCR probes (see online Supplemental Table 4) and detected positive signals in the cfDNA of both CSF and plasma samples (Fig. 3; Table 1). Amplicon sequencing primers targeted to the 8 hotspot mutations were also designed and tested. Four pairs of these primers showing high sensitivity and specificity were used for targeted amplicon sequencing on the cfDNA of CSF and plasma samples (see online Supplemental Table 3).
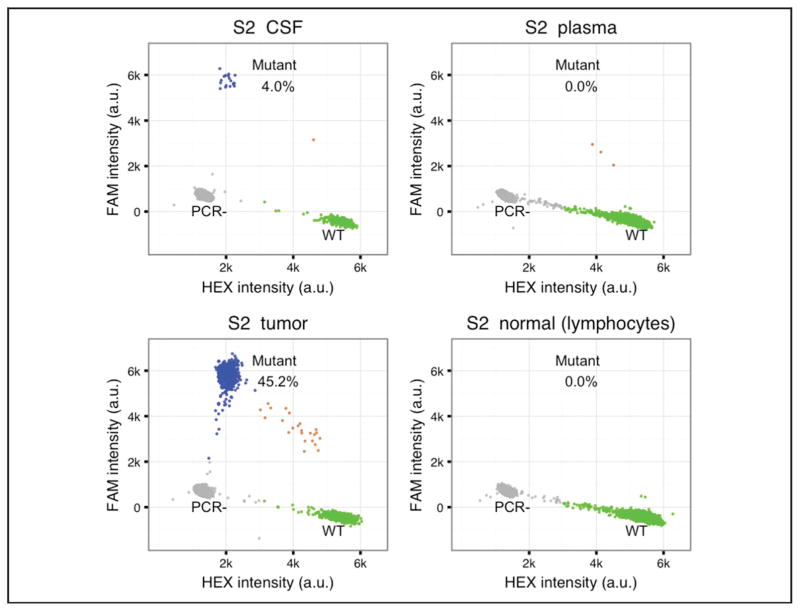
Fig. 3. An example of ddPCR results.
The TaqMan PCR probes were used to detect wild-type (WT) and mutant DNA in CSF, plasma, tumor, and blood cell samples of patient S2. AKT WT probe was labeled with HEX fluorophore and AKT p.E17K probe was labeled with FAM fluorophore. Each dot in the plot represents a droplet. The x axis is HEX fluorescence intensity in arbitrary units (a.u.); the y axis is FAM fluorescence intensity in a.u. Blue, droplets with mutant DNA; green, droplets with wild-type DNA; gray, droplets without target DNA templates; brown, droplets have both mutant and wild-type DNA.
Table 1.
Results of the ddPCR and targeted amplicon sequencing.
| Patient ID | Tumor type | ddPCR measurement
|
AmpSeq measurement
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ddPCR targeted mutation | cfDNA, ng/mL
|
Mutant allele, copies/mL
|
Mutant AF
|
AmpSeq targeted mutation | Mutant AF
|
||||||
| CSF | Plasma | CSF | Plasma | CSF | Plasma | CSF | Plasma | ||||
| S1 | Vestibular schwannoma | NF2 p.R57* | 2.1 | 3.2 | 0 | 0 | 0.000 | 0.000 | NF2 p.R57* | 0.000 | 0.000 |
|
| |||||||||||
| S2 | Atypical meningioma | AKT1 p.E17K | 2.4 | 21.4 | 32 | 0 | 0.040 | 0.000 | AKT1 p.E17K | NP | NP |
|
| |||||||||||
| S3 | Brain metastases from melanoma | BRAF p.V600E | 0.3 | 7.7 | 7 | 0 | 0.074 | 0.000 | BRAF p.V600E | NP | NP |
|
| |||||||||||
| S4 | Brain metastases from melanoma | NRAS p.Q61La | 3.3 | 1.6 | 3 | 117 | 0.003 | 0.214 | NRAS p.Q61L | NP | NP |
|
| |||||||||||
| S5 | Brain metastases from colon adenocarcinoma | KRAS p.G12D | 18.1 | 8.7 | 2963 | 4 | 0.492 | 0.002 | TP53 p.R282W | 0.654 | 0.000 |
|
| |||||||||||
| S6 | Brain metastases from lung adenocarcinoma | EGFR p.L585R | 0.5 | 4.7 | 3 | 693 | 0.020 | 0.445 | EGFR p.L585R | 0.015 | NAb |
|
| |||||||||||
| S7 | Brain metastases from lung adenocarcinoma | EGFR p.E746_A750del | 0.7 | 7.7 | 2 | 0 | 0.009 | 0.000 | EGFR p.E746_A750del | 0.002 | 0.000 |
NRAS, neuroblastoma RAS viral (v-ras) oncogene homolog; NP, no primer available.
NA, not available. The S6 plasma cfDNA sample was not analyzed by targeted amplicon sequencing analysis.
The results of the mutant allele fraction measurements from targeted amplicon sequencing and digital PCR were concordant for both CSF and plasma (Fig. 2C; Table 1). In CSF samples, mutant alleles were detectable for 6 of 7 patients (S1–S7). No mutation was detected in the other sample, which was a grade 1 vestibular schwannoma (a benign tumor of the eighth cranial nerve). The inability to detect tumor mutations in this sample may be due to the anatomic sequestration by arachnoid layers surrounding the tumor. The mutant allele concentration varied between patients, from <5 to nearly 3000 per mL CSF (Fig. 4).
Fig. 4. Quantification of tumor cfDNA in CSF and plasma.
The y axis represents mutant allele concentrations (copies/mL) of both CSF and plasma samples in log10 scale. The demonstrated results were from digital PCR measurement.
COMPARISON OF TUMOR MUTATIONS IN THE cfDNA OF CSF AND PLASMA
For the patients with solid brain tumors (S1–S7), the mutant alleles were quantified in both plasma and CSF cfDNA (Fig. 4; Table 1). For the following discussion, the mutant allele concentration and fraction were based on ddPCR measurements. In 1 primary brain tumor patient (S2, atypical meningioma) and 2 metastatic brain tumor patients (S3, brain metastases from melanoma; S7, brain metastases from lung adenocarcinoma), tumor mutations were not detectable in plasma cfDNA, but the amount of mutant alleles was significant in CSF cfDNA (4% in S2, 7.4% in S3, and 0.9% in S7). In 1 of the metastatic brain tumor patients (S5, brain metastases from colon adenocarcinoma), there was a higher mutant allele fraction in CSF than in plasma. In contrast, the plasma samples from 2 other patients with widely disseminated cancer as documented by positron-emission/computed tomography (S4, brain metastases from melanoma; S6, brain metastases from lung adenocarcinoma) revealed much higher mutant allele fractions in plasma compared to CSF. This higher level of mutant allele fraction in plasma was likely related to the metastatic tumors outside the CNS (S4, metastatic disease in the lungs, skeleton, and abdomen; S6, metastatic disease in the liver). For the patient with a grade I vestibular schwannoma (S1), tumor alleles could not be detected in either CSF or plasma. Overall, these results support our hypothesis that mutations originating in brain tumor are more detectable in CSF than in plasma when the systemic metastatic burden is low.
CHARACTERIZATION OF GENOMIC ABERRATIONS BY CSF CANCER PANEL SEQUENCING
The presence of tumor mutations in the CSF of patients with brain tumors provides an opportunity to diagnose and characterize brain tumor genomic aberrations through CSF DNA analysis. Our technique could potentially be used as an alternative or adjuvant diagnostic assay to the more morbid brain tumor tissue biopsy or insensitive cytology. To test this approach and to eliminate the need for the exome sequencing of tumor/healthy tissue, we applied cancer panel sequencing on CSF total DNA to comprehensively characterize the brain tumor mutation profile. To increase sequencing coverage and reduce cost for eventual use in the clinical setting, our method targeted coding regions of known cancer genes.
For proof-of-principle of the cancer panel sequencing approach, we tested the CSF sample from a patient with suspected leptomeningeal metastasis from lung adenocarcinoma (L1). From the same lumbar puncture required for diagnosis, 10 mL CSF was processed via the cancer panel sequencing method, from which 30 ng total DNA (including both cell-free DNA and cellular genomic DNA) was isolated. Deep sequencing with 160 million reads achieved a coverage of approximately 2000 (before duplication removal). To correct for sequencing error, the reads were redundantly sequenced, and consensus sequences were identified on the basis of collapsing reads with identical genome coordinates (18, 24). The average coverage of the consensus reads was approximately 300.
The primary lung tumor of the same patient had been molecularly characterized in a clinical pathology laboratory with PCR followed by restriction enzyme digestion and capillary electrophoresis fragment analysis. The clinical results showed positive for EGFR (epidermal growth factor receptor)6 c.2573 T>G mutation and negative for KRAS (Kirsten rat sarcoma viral oncogene homolog) codon 12/13 mutations. Our CSF cancer panel sequencing results agreed with these results from the primary tumor sample. We identified 7 somatic SNV mutations (Table 2). Among these mutations, 2 were in well-known cancer genes EGFR and TP53 (tumor protein p53). The first was EGFR c.2573 T>G mutation with the mutant allele fraction of 0.315. The second was TP53 c.338T>G mutation, which has been previously reported as a somatic mutation in lung cancer (25, 26). No mutation was found at KRAS codon 12/13, consistent with the genetic test result of the primary tumor. The patient’s clinical cytologic exam revealed malignant cells in the CSF, confirming the presence of leptomeningeal metastases and concordant with our results.
Table 2.
CSF cancer panel sequencing identifies somatic SNV mutations from the CSF of a patient with leptomeningeal disease (L1).
| Mutation type | Gene | Chromosome | Position | Reference allele | Mutant allele | Mutant AF |
|---|---|---|---|---|---|---|
| Nonsynonymous SNV | EGFR | 7 | 55259515 | T | G | 0.315 |
| Nonsynonymous SNV | TP53 | 17 | 7579349 | A | C | 0.178 |
| Synonymous SNV | MLL2a | 12 | 49444973 | T | A | 0.167 |
| Nonsynonymous SNV | NSD1 | 5 | 176722183 | A | C | 0.083 |
| Nonsynonymous SNV | CREB3L1 | 11 | 46329492 | A | T | 0.076 |
| Nonsynonymous SNV | TPR | 1 | 186324685 | C | A | 0.075 |
| Nonsynonymous SNV | TSC2 | 16 | 2121589 | T | G | 0.072 |
MLL2, •••; NSD1, nuclear receptor binding SET domain protein 1; CREB3L1, cAMP responsive element binding protein 3-like 1; TPR, translocated promoter region, nuclear basket protein; TSC2, tuberous sclerosis 2.
Discussion
In this study, we demonstrated that tumor mutations were detectable in the CSF of patients with different types of brain tumors. To our knowledge, this is the first high-throughput sequencing–based method to characterize brain tumor cfDNA in CSF. We also applied cancer panel sequencing to identify genomic aberrations from CSF total DNA with cancer panel sequencing. Our results demonstrated that cfDNA from brain tumors was present at higher concentrations in CSF than in plasma when the systemic disease burden was low. This implied that CSF cfDNA may be used to detect mutations within CNS malignancies despite low plasma cfDNA concentrations.
We presented 2 strategies to detect tumor mutations in CSF that can be applied to clinical diagnosis. The first strategy, an approach based on digital PCR and targeted amplicon sequencing, is cost effective and highly sensitive. However, it requires either the knowledge of the patient’s tumor mutations before CSF analysis or the use of a panel of frequent mutation sites on the basis of tumor-specific epidemiological studies. For patients who have had tumor resections, their tumor mutations can be identified from the surgical sample with standard clinical tests, such as sequencing or genotyping arrays. Should the patient then require CSF testing to rule out leptomeningeal metastases, the CSF could be analyzed for the known mutations. The tumor types that most commonly metastasize to the brain, such as breast, lung, and melanoma, have well characterized hotspot mutations amenable to this strategy. The second strategy, cancer panel sequencing, could be used as a minimally invasive “liquid biopsy” to analyze the brain tumor genome. Similar to the application of plasma tumor cfDNA (24), this strategy has potential to guide personalized cancer therapy, monitor residual disease, and track the evolving tumor genome.
Supplementary Material
Acknowledgments
We thank Lawrence Shuer, Griffith Harsh, Steven Chang, and Gordon Li for sample collection. We thank Yoon-Jae Cho, Charles Gawad, and John Beausang for helpful discussions. We also thank Norma Neff, Ben Passarelli, Gary Mantalas, and Sean Liu for help in performing sequencing experiments.
Footnotes
Nonstandard abbreviations: cfDNA, cell-free DNA; CNS, central nervous system; CSF, cerebral spinal fluid; ddPCR, droplet digital PCR; SNV, single-nucleotide variant; COSMIC, catalog of somatic mutations in cancer; UID, unique identifier.
Human genes: EGFR, epidermal growth factor receptor; KRAS, Kirsten rat sarcoma viral oncogene homolog; TP53, tumor protein p53; NF2, neurofibromin 2 (merlin).
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures or Potential Conflicts of Interest: No authors declared any potential conflicts of interest.
Role of Sponsor: No sponsor was declared.
References
- 1.Murtaza M, Dawson S-J, Tsui DWY, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–12. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 2.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weston CL, Glantz MJ, Connor JR. Detection of cancer cells in the cerebrospinal fluid: current methods and future directions. Fluids Barriers CNS. 2011;8:14. doi: 10.1186/2045-8118-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi W, Lv C, Qi J, Zhao W, Wu X, Jing R, et al. Prognostic value of free DNA quantification in serum and cerebro-spinal fluid in glioma patients. J Mol Neurosci. 2012;46:470–5. doi: 10.1007/s12031-011-9617-0. [DOI] [PubMed] [Google Scholar]
- 5.Chen WW, Balaj L, Liau LM, Samuels ML, Kotsopoulos SK, Maguire CA, et al. BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol Ther Nucleic Acids. 2013;2:e109. doi: 10.1038/mtna.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhodes CH, Honsinger C, Sorenson GD. PCR-detection of tumor-derived p53 DNA in cerebrospinal fluid. Am J Clin Pathol. 1995;103:404–8. doi: 10.1093/ajcp/103.4.404. [DOI] [PubMed] [Google Scholar]
- 7.Swinkels DW, de Kok JB, Hanselaar A, Lamers K, Boerman RH. Early detection of leptomeningeal metastasis by PCR examination of tumor-derived K-ras DNA in cerebrospinal fluid. Clin Chem. 2000;46:132–3. [PubMed] [Google Scholar]
- 8.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A. 1999;96:9236–41. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu W, Koh W, Blumenfeld YJ, El-Sayed YY, Hudgins L, Hintz SR, et al. Noninvasive prenatal diagnosis in a fetus at risk for methylmalonic acidemia. Genet Med. 2014;16:564–7. doi: 10.1038/gim.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, et al. The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin Chem. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- 17.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3: new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–5. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinde I, Bettegowda C, Wang Y, Wu J, Agrawal N, Shih I-M, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med. 2013;5:167ra4. doi: 10.1126/scitranslmed.3004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glass JP, Melamed M, Chernik NL, Posner JB. Malignant cells in cerebrospinal fluid (CSF): the meaning of a positive CSF cytology. Neurology. 1979;29:1369–75. doi: 10.1212/wnl.29.10.1369. [DOI] [PubMed] [Google Scholar]
- 21.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deisenhammer F, Bartos A, Egg R, Gilhus N, Giovannoni G, Rauer S, et al. Routine cerebrospinal fluid (CSF) analysis. In: Gilhus NE, Barnes MR, Brainin M, editors. European handbook of neurologic management. New York: John Wiley and Sons; 2011. pp. 5–17. [Google Scholar]
- 23.Pati N, Schowinsky V, Kokanovic O, Magnuson V, Ghosh S. A comparison between SNaPshot, pyrosequencing, and biplex invader SNP genotyping methods: accuracy, cost, and throughput. J Biochem Biophys Methods. 2004;60:1–12. doi: 10.1016/j.jbbm.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Newman AM, Bratman SV, To J, Wynne JF, Eclov NCW, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–54. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seo J-S, Ju YS, Lee W-C, Shin J-Y, Lee JK, Bleazard T, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–19. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casey G, Lopez ME, Ramos JC, Plummer SJ, Arboleda MJ, Shaughnessy M, et al. DNA sequence analysis of exons 2 through 11 and immunohistochemical staining are required to detect all known p53 alterations in human malignancies. Oncogene. 1996;13:1971–81. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.