König et al. perform time-resolved electron tomographic reconstructions to decipher the membrane remodeling events in abscission, the final step of cytokinesis, in Caenorhabditis elegans and find that completion of the scission process requires actomyosin-driven membrane remodeling, but not the ESCRT machinery.
Abstract
Abscission is the final step of cytokinesis and results in the physical separation of two daughter cells. In this study, we conducted a time-resolved series of electron tomographic reconstructions to define the steps required for the first embryonic abscission in Caenorhabditis elegans. Our findings indicate that membrane scission occurs on both sides of the midbody ring with random order and that completion of the scission process requires actomyosin-driven membrane remodeling, but not microtubules. Moreover, continuous membrane removal predominates during the late stages of cytokinesis, mediated by both dynamin and the ESCRT (endosomal sorting complex required for transport) machinery. Surprisingly, in the absence of ESCRT function in C. elegans, cytokinetic abscission is delayed but can be completed, suggesting the existence of parallel membrane-reorganizing pathways that cooperatively enable the efficient severing of cytoplasmic connections between dividing daughter cells.
Introduction
In metazoan cells, cytokinesis is initiated by signals from the anaphase spindle, triggering the assembly of an actomyosin ring that constricts the plasma membrane to generate a narrow intercellular bridge containing two bundles of antiparallel microtubules that overlap in the central zone called the midbody (Glotzer, 2005; Green et al., 2012). The intercellular bridge is subsequently severed on both sides of the midbody at secondary constrictions, and the midbody remnant is released and subsequently engulfed by one of the daughter cells (Ettinger et al., 2011; Chai et al., 2012; Agromayor and Martin-Serrano, 2013; Elia et al., 2013; Lafaurie-Janvore et al., 2013; Crowell et al., 2014). Several models have been developed to describe the mechanism of abscission (Schiel and Prekeris, 2010; Mierzwa and Gerlich, 2014; Gould, 2016; Schöneberg et al., 2017). Although previous studies suggest that internal membrane compartments fill the intercellular channel and template membrane fusion reactions that divide the cell (Finger and White, 2002; Xu et al., 2002; Low et al., 2003), more recent evidence suggests a role for Rab11/FIP3-positive recycling endosomes and Rab35-bound vesicles in delivering key factors, including OCRL, SCAMP2/3, and p50RhoGAP, which regulate actin organization and help to establish secondary constrictions at future abscission sites (Kouranti et al., 2006; Dambournet et al., 2011; Schiel et al., 2012). Microtubules play an essential role in this process by serving as tracks along which vesicles are transported toward constriction sites, but they must be depolymerized before membrane scission, which is thought to be mediated by the ESCRT (endosomal sorting complex required for transport) machinery (Caballe and Martin-Serrano, 2011; Guizetti et al., 2011; Schiel et al., 2011; Mierzwa and Gerlich, 2014; Hurley, 2015; Schöneberg et al., 2017). Specifically, ESCRT-III filaments exhibit the ability to remodel lipid bilayers, facilitating membrane constriction and fission (Elia et al., 2012; McCullough et al., 2013; Schuh and Audhya, 2014; Hurley, 2015; Schöneberg et al., 2017). In general, current models on cytokinesis are based on cytological changes observed in cultured mammalian cells grown on artificial substrates, begging the question of what actually occurs in an in vivo setting.
The Caenorhabditis elegans early embryo offers several advantages to study abscission in a native setting. First, embryo development is highly stereotyped, enabling accurate correlation of live-cell imaging studies with ultrastructural analysis of cryoimmobilized cells (Pelletier et al., 2006). Second, the reproducible timing and duration of cytokinesis in the one-cell C. elegans embryo also permits definition of the acute impacts of disrupting individual cytoskeletal elements at various time points during furrow ingression (Oegema and Hyman, 2006). Third, the C. elegans germline is particularly sensitive to protein depletion after RNAi, allowing identification of the roles of specific factors implicated in cytokinesis during the first time abscission takes place (Oegema and Hyman, 2006). This contrasts with depletion studies in mammalian tissue culture cells, in which protein elimination requires multiple days and cells continue to divide, potentially in an aberrant manner, under conditions in which protein levels are partially reduced.
Previous work using the early C. elegans embryo has helped to define discrete steps that take place during cytokinesis (Green et al., 2013). However, a high-resolution ultrastructural view of these events has been lacking. To understand how the topology of the intercellular bridge evolves during cell division, we used a correlative light and electron microscopic approach. Our findings highlight conserved mechanisms that contribute to the abscission process but also demonstrate unique features that distinguish cytokinesis in an embryonic system from that observed in mammalian cells in culture.
Results and discussion
Ultrastructural analysis of the first embryonic cytokinesis in C. elegans
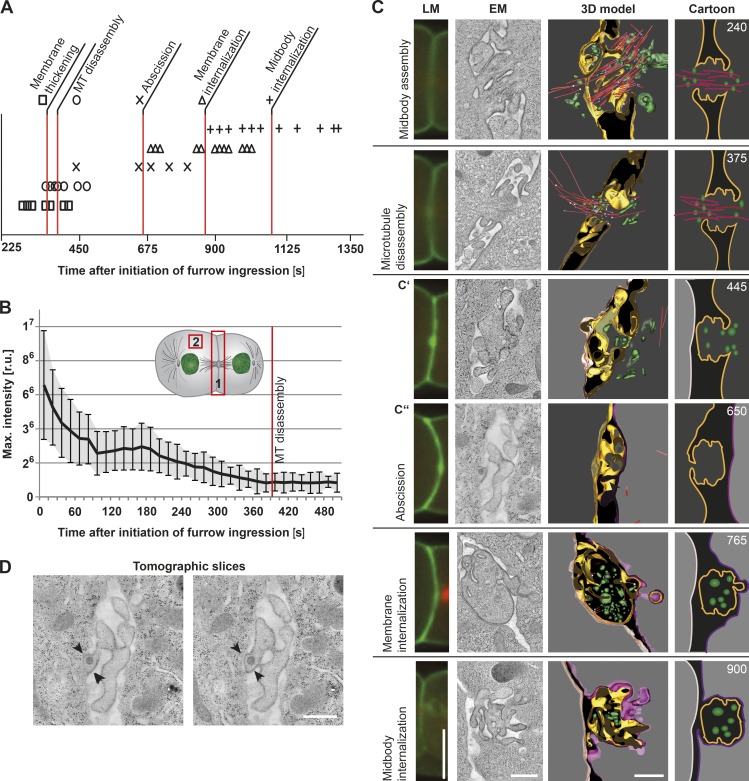
To characterize the first embryonic cell division in C. elegans, we initially used spinning-disk confocal microscopy and conducted time-lapse imaging of transgenic zygotes coexpressing probes for the plasma membrane (GFP::PH [pleckstrin homology]) and microtubules (mCherry::β-tubulin). These studies reproducibly demonstrated the timing of various events relative to the onset of furrow ingression. In particular, we found that completion of furrow ingression occurred within 170 s (± 28 s; n = 11; Fig. S1 A). Thickening of the plasma membrane followed (360 ± 49 s; n = 13) and was accompanied by microtubule disassembly shortly thereafter (389 ± 45 s; n = 15), as indicated by a local increase in the intensity of the GFP::PH probe and a loss in mCherry::β-tubulin fluorescence, respectively (Fig. 1, A and B; and Fig. S1 A). We also consistently observed the asymmetric internalization of GFP::PH-labeled membranes into the cytoplasm of the posterior cell (876 ± 117 s; n = 12; Fig. 1 A). Finally, complete engulfment of the midbody into the posterior cell occurred ∼1,088 s after the onset of furrow ingression (± 137 s; n = 13; Fig. 1 A).
Figure 1.
Time-resolved analysis of cytokinesis by correlative light and electron microscopy. (A) Timing of abscission events during the first embryonic division. The start of cleavage furrow ingression is defined as the 0 s time point. Single time points for each analyzed abscission event are indicated by symbols. A red line highlights the mean time point for each. The time point of abscission was obtained by tomographic analysis as shown in C. n = 11–15, depending on the step analyzed. (B) Determination of the time point of microtubule disassembly. Analysis of fluorescence intensity (mCherry::β-tubulin) in the region of the cleavage furrow (position 1) and in the cytoplasm (defined as background; position 2). The background fluorescence was subtracted from the fluorescence intensity at the cleavage furrow. The mean values of 10 single embryos were plotted over time. The time point of microtubule (MT) disassembly is indicated (red line). Error bars show SD. r.u., relative units. (C) 3D reconstruction of intercellular bridges at defined time points after initiation of furrow ingression as indicated in the right panels (top right corner, seconds). A light microscopy (LM) image (first column, GFP::PH in green and mCherry::β-tubulin in red), an electron microscopy image (second column, tomographic slice), the corresponding 3D model (third column), and a cartoon illustration (fourth column) are shown for each time point. Color coding is as follows: gold, plasma membrane of an open side of the intercellular bridge; red, microtubules; white, open microtubule ends; blue, closed microtubule ends; green, vesicles; dark gray background, cytoplasm at the open bridge; light gray background, cytoplasm at the disconnected bridge; nude, anterior cell membrane; and purple, posterior cell membrane. The midbody remnants (765 s and 900 s) are also shown in gold. (D) Additional tomographic z slices of an embryonic intercellular bridge (650 s; a thickening of the membrane is indicted by arrowheads). Bars, 500 nm.
Previous work defined abscission as a two-step process involving cytoplasmic isolation between daughter cells followed by midbody release (Green et al., 2013). However, this work relied largely on light microscopy to define stages of abscission, which provides insufficient resolution to determine changes in membrane topology and cytoskeletal organization during each stage of cytokinesis. We therefore performed correlative light and electron microscopy on isolated embryos that were high-pressure frozen at specific time points after furrow ingression began, and then we collected electron tomograms from serial semithick sections (Fig. 1 C and Videos 1, 2, 3, 4, 5, and 6). A representative early intercellular bridge, frozen 240 s after onset of furrow ingression, was 0.75 µm in length, 1.5 µm in diameter, and contained an array of midzone microtubules. Electron tomograms also revealed an accumulation of vesicles, which were on average 55 ± 4 nm in diameter and distributed throughout the intercellular bridge, as well as tubulovesicular organelles in the surrounding area and numerous membrane ridges protruding from the midbody. In many ways, the structure of the C. elegans intercellular bridge at this time point resembles that of a HeLa cell in early telophase (Schiel et al., 2011). At a later time point before abscission (375 s after initiation of furrow ingression), microtubules exhibited a more asymmetric distribution (Fig. 1 C). In some cases, the majority of the microtubules originated in the anterior cell and extended into the intercellular bridge, whereas in other cases, the posterior cell exhibited a higher density of microtubules. The positioning of vesicles within the region mirrored the asymmetry in microtubules, suggesting that they arrive using microtubule tracks and contribute to the delivery of vesicle-associated signaling factors during the early stages of cytokinesis before abscission. To test this idea, we depleted animals of PERM-1 (to permeabilize the embryonic eggshell) and treated one cell–stage embryos with the microtubule-depolymerizing drug nocodazole upon initiation of furrow ingression. Under conditions where microtubules were absent within the intercellular bridge based on both light and electron microscopy, we failed to observe an accumulation of membranes near the midbody, and the intercellular bridge exhibited a reduced diameter (350 nm) in comparison with control embryos (∼240 s after onset of furrow ingression; Fig. S1, B and C). Nonetheless, abscission was able to complete under these conditions, consistent with prior work both in C. elegans and mammalian cells (Guizetti et al., 2011; Schiel et al., 2011; Green et al., 2013). In the absence of nocodazole, PERM-1–depleted embryos exhibited normal timing of all events associated with abscission (Fig. S1 D).
We next used electron tomography to define the ultrastructure of the intercellular bridge during abscission, analyzing serial semithick sections to ensure that small connections between daughter cells were not overlooked. These data demonstrated that membrane sealing around the midbody occurred between 450 and 850 s after initiation of furrow ingression (Fig. 1, A and C), subsequent to cytoplasmic isolation but well before midbody release, which was previously suggested to correlate with abscission (Green et al., 2013). At no point during this time period did we observe microtubules within the intercellular bridge (Fig. 1 C). In addition, we analyzed multiple cases in which one of the daughter cells had been abscised, whereas the other daughter cell remained connected to the intercellular bridge. In some instances, the anterior cell underwent abscission before the posterior cell (four out of eight embryos; Fig. 1 C′, 445 s), but in others, the posterior cell was abscised first (four out of eight embryos; Fig. 1 C′′, 650 s). Our findings suggest that the first abscission event occurs randomly and without preference for one cell over another. Additionally, to our surprise, we did not detect helical filaments or other abscission-specific structures identified previously in mammalian tissue culture cells (Guizetti et al., 2011) in any serial section during the time period of abscission in C. elegans embryos (Fig. 1 D).
After completion of plasma membrane sealing, the residual body of the intercellular bridge, termed the midbody remnant, remained between the daughter cells (Fig. 1 C, 765 s). At this stage, we observed small membrane buds emanating from the midbody remnant, which appeared to be undergoing engulfment into the cytoplasm of the posterior cell (Fig. 1 C). The appearance of these nascent membrane budding structures correlated with the time point of GFP::PH-labeled membrane internalization observed using light microscopy. Approximately 900 s after the onset of furrow ingression, 60% of the embryos (n = 15) displayed a midbody remnant that was completely enclosed by the plasma membrane and engulfed or actively undergoing engulfment into the posterior cell (Fig. 1 C).
The ESCRT machinery is required for membrane internalization after abscission
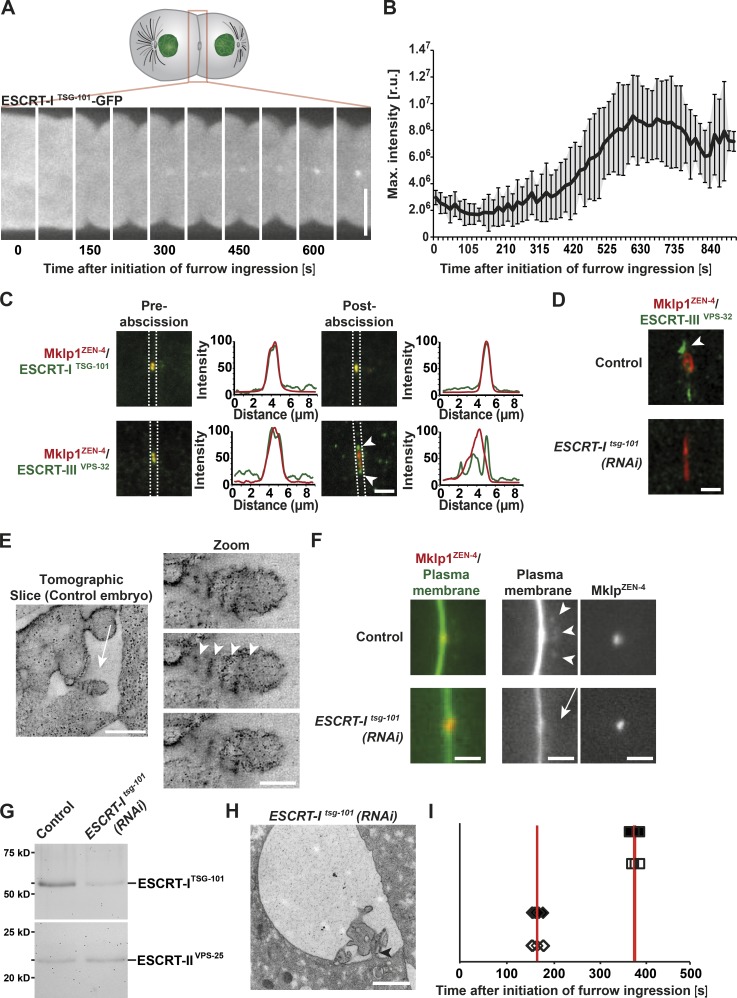
Although initially described for their role in endosomal cargo sorting, components of the ESCRT machinery have been implicated in a wide variety of membrane remodeling events, including those that occur during cytokinesis (Schuh and Audhya, 2014; Hurley, 2015; Christ et al., 2017). However, the precise role for the ESCRTs during abscission remains controversial. To address this issue, we first examined the distribution of the ESCRT-I subunit TSG-101 during embryonic cell division in C. elegans. Initially, TSG-101 localized to the mitotic spindle and was subsequently detected at the intercellular bridge ∼300 s after initiation of furrow ingression (Fig. 2 A). Accumulation of TSG-101 plateaued ∼300 s later and remained constant at the midbody thereafter (Fig. 2 B). Consistent with these findings, endogenous TSG-101 colocalized with the midbody component ZEN-4 both before and after abscission (Fig. 2 C). In contrast, the ESCRT-III component VPS-32 was distributed largely at the edges of the midbody remnant at these time points in a manner dependent on the presence of TSG-101 (Fig. 2, C and D).
Figure 2.
The ESCRT machinery promotes membrane internalization after abscission. (A) Representative images of ESCRT-ITSG-101 accumulation at the intercellular bridge by fluorescence light microscopy. Bar, 10 µm. (B) Quantification of ESCRT-ITSG-101 accumulation. Error bars show SD. r.u., relative units. n = 10. (C) Confocal images showing the accumulation of MKLP1ZEN-4 together with ESCRT-ITSG-101 or ESCRT-IIIVPS-32 before and after abscission. Arrowheads indicate the distinct accumulation of MKLP1ZEN-4 and ESCRT-IIIVPS-32 after abscission. Line scan analysis within the boxed regions was used to measure the distributions of ESCRT components relative to ZEN-4 in each image. n = 6 embryos for each condition. Bar, 2 µm. (D) STED microscopy indicating the accumulation of ESCRT-IIIVPS-32 in control (arrowhead) and tsg-101(RNAi) embryos. n = 6 embryos for each condition. Bar, 1 µm. (E) Tomographic slice of a midbody remnant showing membrane internalization (left, arrow; bar, 500 nm) and a series of tomographic slices of the same membrane bud (right; bar, 100 nm). The spacing of the filaments is indicated (arrowheads). (F) Fluorescence light microscopy of the cleavage furrow of a control (top) and a tsg-101(RNAi) embryo (bottom). Membranes and midbodies were visualized by expressing GFP::PH and mCherry-MKLP1ZEN-4, respectively. Membrane internalization (arrowheads) is visible in the control but absent in the tsg-101(RNAi) embryo (arrow). Bars, 2 µm. n = 12. (G) Immunoblot analysis of control and TSG-101–depleted whole animals using antibodies directed against TSG-101 and VPS-25 (Schuh et al., 2015), a component of the downstream ESCRT-II complex. n = 3. (H) Representative electron micrograph of a midbody remnant in a tsg-101(RNAi) embryo taken 1,100 s after initiation of furrow ingression. The connection of the intercellular bridge to one of the daughter cells is indicated (arrowhead). Bar, 1 µm. n = 3 embryos. (I) Timing of abscission events during the first embryonic division in control and TSG-101–depleted embryos. The start of cleavage furrow ingression is defined as the 0 s time point. Cleavage furrow closure (open diamonds, control; closed diamonds, TSG-101–depleted embryos) and microtubule disassembly (open squares, control; closed squares, TSG-101–depleted embryos) are shown. Red lines highlight the mean time point for each. n = 6 embryos for each condition.
In vitro analysis previously demonstrated that recombinant C. elegans VPS-32 assembles into spiral filaments with a thickness of 4.2 nm (Shen et al., 2014). To determine whether the accumulation of VPS-32 near the midbody correlated with the presence of spiral filaments as observed previously in mammalian cells undergoing abscission (Guizetti et al., 2011), we again took advantage of electron tomography. Although we failed to observe filamentous structures throughout the abscission process (Fig. 1, C and D), we consistently identified filamentous structures at the necks of membrane buds emerging from the midbody remnant ∼800 s after the onset of furrow ingression (Fig. 2 E). These membrane bud–associated filaments exhibited a diameter of 12 nm and a spacing of 26 nm, slightly thicker than the recombinant VPS-32 spiral polymers observed by cryo-electron microscopy (Shen et al., 2014) but consistent with the idea that these filaments correspond to ESCRT-III heteropolymers. Upon depletion of TSG-101, which inhibits downstream recruitment of the ESCRT-III complex (Fig. 2 D), GFP::PH-labeled membrane internalization into the posterior cell was no longer observed (12/12 control embryos exhibited membrane internalization, whereas 0/12 tsg-101(RNAi) embryos showed evidence of membrane internalization; Fig. 2 F), arguing that the ESCRT machinery plays a key role in this process. Our findings are in contrast to those observed after Shrub (VPS-32) depletion in Drosophila melanogaster S2 cells, in which midbody membrane shedding continued to occur in the absence of normal ESCRT-III assembly (El Amine et al., 2013). Although it is possible that ESCRT-I activity at the midbody extends beyond that of ESCRT-III recruitment, an alternative explanation is that we were able to achieve more penetrant depletion in C. elegans embryos (on average >90% depletion of TSG-101 based on whole-animal immunoblots; Fig. 2 G).
To more closely examine the impact of inhibiting ESCRT recruitment to the midbody, we froze embryos depleted of TSG-101 ∼1,100 s after initiation of furrow ingression, a time point after midbody remnant engulfment in control embryos. In parallel, we verified that ESCRT-dependent cargo degradation was strongly inhibited, ensuring penetrant protein depletion had been achieved (Audhya et al., 2007). In agreement with previous fluorescence-based imaging analysis (Green et al., 2013), we found that the midbody remnant remained trapped between daughter cells under these conditions (Fig. 2 H). Moreover, with the improved resolution afforded by serial thin-section electron microscopy, we were able to confirm the absence of membrane budding from the midbody, and we failed to observe assembly of any membrane-associated spiral filaments (Figs. 2 H and S2, A and B). Instead, the membrane surface area surrounding the remnant became swollen, consistent with the idea that the ESCRT machinery normally acts to facilitate membrane removal during the abscission phase of cell division. Surprisingly, we also found that the intercellular bridge on one side of the midbody was consistently severed, whereas thin cytoplasmic connections remained on the opposite side (Figs. 2 H and S2 and B). Although RNAi-mediated depletion of TSG-101 is unlikely to achieve 100% protein elimination, these data suggest that cytokinetic abscission can occur in the absence of ESCRT function, although the process appears to be delayed. To determine whether ESCRT function may be required during earlier phases of cytokinesis and thereby affects the timing of abscission, we depleted TSG-101 and defined its impact on the rate of furrow ingression and on the timing of microtubule disassembly. Our studies revealed that ESCRT function is dispensable for both processes (Fig. 2 I), consistent with the relatively late accumulation of TSG-101 at the midbody during cytokinesis. Collectively, these data suggest a new role for ESCRTs in membrane removal during the final steps of cell division, a conserved process that has also been observed in other metazoan systems (El Amine et al., 2013; Sherman et al., 2016).
Actin polymers play an active role during the late stages of abscission
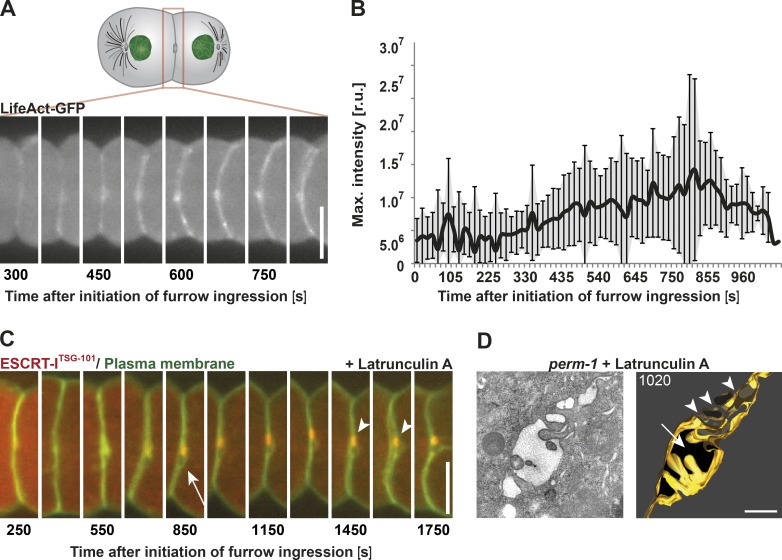
Although actin plays a well-described role in driving cortical contractility during cytokinesis (Mishima, 2016), its roles during late stages of abscission remain unclear (Guizetti et al., 2011; Prekeris, 2011; Schiel et al., 2012; Mierzwa and Gerlich, 2014). Although several lines of evidence indicate that actin is required for initial stabilization of the intercellular bridge, actin filaments must ultimately be cleared for the final abscission step to take place (Dambournet et al., 2011; Schiel et al., 2012). Using transgenic embryos expressing LifeAct-GFP to visualize actin filaments during cytokinesis, we observed an initial signal at the intercellular bridge ∼375 s after initiation of furrow ingression (Fig. 3 A; Singh and Pohl, 2014), and fluorescence intensity of the probe peaked ∼425 s later (Fig. 3 B). To define the role of actin during this time interval, we permeabilized the embryonic eggshell using PERM-1 depletion and treated one cell–stage embryos with latrunculin A upon furrow closure, after intercellular bridge assembly was complete (∼170 s after initiation of furrow ingression; Fig. 3 C). The embryos accumulated TSG-101 at the midbody normally, but neither membrane internalization nor engulfment of the midbody remnant was detected (Fig. 3 C, arrowheads). Additionally, we observed a persistent membrane opening adjacent to the midbody ∼550 s after onset of furrow ingression in 80% of the embryos (∼2.8 µm in length and 1.5 µm in width on average; n = 15; Fig. 3 C, white arrow). These data suggest that actin polymers play a role late during the abscission process.
Figure 3.
Polymerized actin is required to complete cytokinetic abscission. (A) Representative still images taken of a single embryo show the accumulation of actin at the intercellular bridge by fluorescence light microscopy using a LifeAct-GFP probe. (B) Quantification of actin accumulation after onset of furrow ingression. Error bars show SD. r.u., relative units. n = 15. (C) Representative still images of a latrunculin A–treated embryo during the late stages of cytokinesis. The drug was added after completion of furrow ingression. Localization of the midbody marker ESCRT-ITSG-101 is indicated (arrowheads). A membrane opening (arrow) is visible next to the midbody. n = 12/15. (D) Tomographic slice (left) and 3D model (right) of a high-pressure frozen latrunculin A–treated embryo. The membrane opening next to the intercellular bridge and a fragmentation of the midbody are indicated by an arrow and arrowheads, respectively. The membrane of the intercellular bridge is highlighted in gold. Bars: (A and C) 10 µm; (D) 500 nm.
To further explore this phenotype, we analyzed the ultrastructure of intercellular bridges in latrunculin A–treated embryos, frozen ∼1,020 s after onset of furrow ingression and long after abscission normally completes (Fig. 3 D). Under this condition, serial semithick section electron tomography revealed that depletion of actin polymers prevents abscission. Instead, the intercellular bridge appeared fragmented (Fig. 3 D, arrowheads) and exhibited multiple thin cytoplasmic connections bridging the embryonic daughter cells. In addition, the membrane opening as visualized by light microscopy could be detected (Fig. 3, C and D, arrows). We observed a similar phenotype by treating permeabilized embryos with Blebbistatin, an inhibitor of myosin II, after completion of furrow ingression (∼170 s after onset of furrow ingression; Fig. S3 A). Specifically, serial semithick section electron tomography of a Blebbistatin-treated embryo frozen ∼1,100 s after onset of furrow ingression showed an intercellular bridge with thin cytoplasmic connections (Fig. S3 B). A previous fluorescence-based imaging study using a temperature-sensitive allele suggested that myosin II is dispensable after completion of furrow ingression (Davies et al., 2014), but our studies provide evidence to the contrary, again highlighting the importance of conducting correlative electron microscopy studies. The specific roles of actin and myosin in executing the final step of cell cleavage are currently unclear. However, this pathway may represent a redundant mechanism to ESCRT-mediated membrane abscission in embryonic systems. Alternatively, because a primary function of actin is to stabilize the cleavage furrow, fragmentation of the intercellular bridge upon actin depolymerization may have indirect consequences on membrane remodeling or positioning required for final membrane scission.
Dynamin-mediated endocytosis is required before abscission
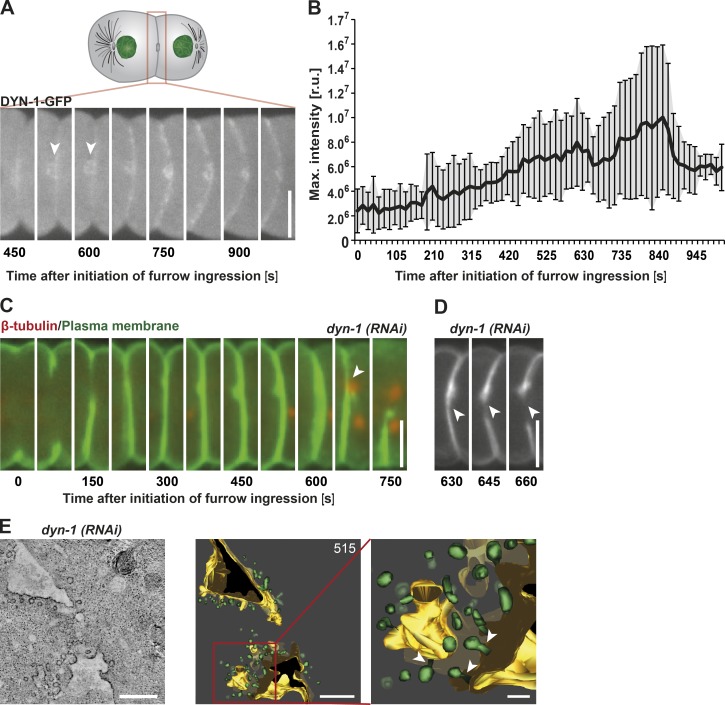
Based on our finding that the ESCRT machinery plays a key role in membrane removal during the late stages of cell division, we decided to explore the impact of eliminating another essential regulator of membrane internalization during embryonic abscission. The large GTPase dynamin, which is encoded by DYN-1 in C. elegans, functions in the severing of nascent endocytic vesicles to generate free transport carriers (Ferguson and De Camilli, 2012). Although implicated in cytokinesis, its precise function during cell division remains unclear. Consistent with previous work (Thompson et al., 2002), we demonstrated that DYN-1 initially accumulates at the intercellular bridge ∼525 s after onset of furrow ingression (Fig. 4 A, arrowheads). Fluorescence intensity measurements of DYN-1::GFP showed a linear accumulation of the protein with a peak ∼350 s later (Fig. 4 B). To define the function of dynamin at the intercellular bridge, we conducted a series of depletion experiments. Midzone architecture in DYN-1–depleted embryos was indistinguishable from control embryos throughout furrow closure, microtubule disassembly, and membrane thickening. However, ∼602 s (± 36 s; n = 5) after onset of furrow ingression, the furrow began to regress (n = 5; Fig. 4, C and D, arrowheads). Based on our finding that myosin levels remained normal on the ingressing furrow in DYN-1–depleted embryos (Fig. S3, C and D), this phenotype was unlikely to be a result of a defect in actomyosin ring stability, as was observed previously in Dictyostelium discoideum after dynamin inhibition (Masud Rana et al., 2013).
Figure 4.
DYN-1 function is required during a late stage of cytokinesis. (A) Representative still images taken of a single embryo show accumulation of dynamin at the intercellular bridge (arrowheads) by fluorescence light microscopy. (B) Quantification of dynamin accumulation after onset of furrow initiation. Error bars show SD. r.u., relative units. n = 10. (C) Representative still fluorescence images of a dyn-1(RNAi) embryo during the late stages of cytokinesis. The membrane is shown in green (GFP::PH) and the microtubules in red (mCherry::β-tubulin). Cleavage furrow regression is indicated (arrowhead). n = 5. (D) Representative still images of a dyn-1(RNAi) embryo showing the start of furrow regression adjacent to the midbody (arrowheads). n = 5. (E) Electron tomography of a dyn-1(RNAi) embryo. The ultrastructure of the intercellular bridge is illustrated in a tomographic slice (left). The corresponding 3D model (middle) and an enlarged region (right) shows an accumulation of nascent vesicles on the plasma membrane (arrowheads). 515 is the time point at which the embryo was frozen and imaged. Bars: (A, C, and D) 10 µm; (E, left and middle) 500 nm; (E, right) 200 nm.
To analyze membrane topology immediately before furrow regression, we froze DYN-1–depleted embryos ∼515 s after onset of cleavage furrow ingression and examined them using electron tomography (Fig. 4 E). These studies revealed broad openings in the daughter cells with diameters of ∼1.25 µm in the anterior cell and ∼0.48 µm in the posterior cell. Both openings appeared to be larger than any observed in control embryos at this time point. Interestingly, membrane ridges protruding from the midbody could not be identified in DYN-1–depleted embryos. However, we observed an accumulation of nascent vesicles on the plasma membrane, consistent with the function of dynamin in releasing endocytic transport carriers as well as free vesicles within the intercellular bridge (Fig. 4 E). These data suggest that DYN-1 plays a key role before abscission to clear excess membrane, which may otherwise interfere with generating a membrane topology conducive to abscission.
Collectively, our findings support a model in which actomyosin-based contractility not only supports cleavage furrow ingression, but also stabilizes the intercellular bridge to appropriately position the juxtaposed plasma membranes before abscission. Surprisingly, scission of the intercellular bridge after the first mitotic division in C. elegans does not appear to exhibit an absolute requirement for the ESCRT machinery, arguing instead for the existence of a parallel, potentially actin-dependent pathway that supports this process. Our findings also highlight the substantial amount of membrane that is transported to the furrow during early embryonic divisions, consistent with previous work examining the first cytokinesis of sea urchin embryos (König et al., 2017). In contrast, cell division in mammalian tissue culture cells involves relatively little change in total plasma membrane surface area (Figard and Sokac, 2014), which may help to explain some of the differences in cytokinesis observed in vitro as compared with embryonic divisions in vivo. Importantly, our data argue that excess membrane must be removed during multiple stages of cell division for the process to proceed normally both in a dynamin-dependent manner and also via function of the ESCRT machinery. Recent work in mammalian cells also described the formation of small membrane blebs near the intercellular bridge during cytokinesis (Sherman et al., 2016), suggesting that this requirement is not unique to worm embryos. In the absence of ESCRT-dependent membrane removal, the membrane surrounding the midbody remnant expands, potentially altering membrane tension, which was shown previously to play an important role in the timing of abscission (Lafaurie-Janvore et al., 2013). In addition, changes in membrane tension may disrupt normal phagocytosis of the midbody remnant into the posterior cell (Masters et al., 2013; Figard and Sokac, 2014). How membrane removal orchestrated by the ESCRT machinery directly contributes to the timing of abscission and midbody internalization represents an important issue to be addressed in the future.
Materials and methods
C. elegans strains
The following strains were used in this study: TH155 (unc-119(ed3)-III; ltIs38[pAA1; pie-1p::GFP::PH(PLC1Δ1);unc-119+]; mCherry::β-tubulin); MSN707 (unc-119(ed3)-III; HzIs169[pie-1p-mCherry::ZEN-4; unc-119+]; ltIs38[pAA1; pie-1p::GFP::PH(PLC1Δ1); unc-119+]); TMR08 (unc-119(ed3)-III; ddIs186[WRM0631C_D06::S000126_R6K-pCFJ496-000032-mos1([240][31816]tsg-101::S000138_R6K-2×TY1-wSNAP-eGFP 3×FLAG); unc-119+]; ltIs44[pAA173; pie-1p-mCherry::PH(PLC1Δ1) + unc-119+]); TMR20 (unc-119(ed3)-III; ddIs86[pie-1p::LifeAct::GFP; unc-119+]; ltIs44[pAA173; pie-1p-mCherry::PH(PLC1Δ1) + unc-119+]); TMR22 (dqls3[pie-1p::dyn1B::GFP + unc-119+]; ltIs44[pAA173; pie-1p-mCherry::PH(PLC1Δ1) + unc-119+]); and MSN608 (unc-119(ed3)-III; HzIs100[pie-1p-GFP::CAV-1; unc-119+]; eri-1(mg366)). Maintenance of C. elegans strains was performed according to standard procedures (Brenner, 1974). For all experiments, L4 larvae were shifted to 25°C 24 h before imaging. For all ultrastructural studies, either strain TH155 or MSN608 was used for staging of early embryos before cryoimmobilization for electron microscopy.
RNAi
RNAi by feeding was performed as described previously (Timmons and Fire, 1998). In brief, L4 larvae or young adult animals were seeded onto bacteria, which had been induced to express specific double-stranded RNAs using IPTG. The following feeding clones were obtained from the Ahringer laboratory and grown at 25°C on feeding plates: T01H3.4 (perm-1; 20–24 h), C09G12.9 (tsg-101; 48 h), and C2C6.1a (dyn-1; 12 h). Using oligonucleotides containing T3 and T7 promotors, double-stranded RNAs were also prepared to deplete TSG-101 by injection: oligonucleotide 1, 5′-TAATACGACTCACTATAGGGGCAAGTGTTATTTGGCGTT-3′, and oligonucleotide 2, 5′-AATTAACCCTCACTAAAGGTATCCAATGGGAGCTTCTGG-3′. Regions of TSG-101 were amplified by PCR using N2 genomic DNA as a template. PCR samples were purified using the PCR cleanup kit (QIAGEN) and eluted with 50 µl of water. The Ambion kit was used for the T3 and T7 transcription reactions. The reaction mix was incubated at 37°C for 5 h. To remove the DNA template, 1.3 µl DNase was added to each reaction and incubated for 15 min at 37°C. The samples were then purified using the RNeasy kit (QIAGEN). Each reaction was eluted with 30 µl RNase-free water. To anneal the RNAs, equal amounts of the T3 and T7 reactions were combined and incubated at 68°C for 10 min and then at 37°C for 1 h. To ensure full depletion of TSG-101 for electron microscopic studies, an eri-1(mg366) mutant background was used, which exhibits hypersensitivity to RNAi (Kennedy et al., 2004).
Live-cell imaging
For live-cell imaging, adult hermaphrodites were transferred to a droplet of M9 buffer and placed on a 20 × 64–mm high-precision cover glass (Roth). Hermaphrodites were dissected to release embryos. Imaging at 20°C was performed on a spinning-disk microscope (ZEISS) using a C-Apochromat 63×/1.2-W objective lens and a CSU-X1 scan head (Yokogawa Electric Corporation). The 405-, 488-, and 561-nm lasers were used for illumination. For time-lapse acquisitions, a double beam splitter 527/54 + 645/60 was used in front of an electron-multiplying charge-coupled device camera (Evolve 515; Roper Technologies). Images were acquired using ZenBlue software (ZEISS) and processed with FIJI (ImageJ; National Institutes of Health; Walter et al., 2010). For quantification of membrane internalization, a threshold was established to identify GFP::PH-labeled membranes that were internalized in control embryos. After depletion of TSG-101, the same threshold was used, and we were unable to observe membrane internalization.
Permeabilization of embryos and drug treatment
To perform drug treatment experiments, eggshells of early embryos were permeabilized by perm-1(RNAi) treatment (Carvalho et al., 2011). Permeabilized embryos were gently aspirated into cellulose capillaries to allow a transfer of embryos into the medium containing the drug (König et al., 2017). Embryos were initially imaged without addition of a drug but in the presence of 2 µg/ml Hoechst to verify the permeability of the eggshell of each selected embryo. At the time point of interest, the capillary with the enclosed embryo was transferred to a new coverslip with media containing the drug, and imaging was continued. The following drugs were added: 10 µM latrunculin A after completion of furrow ingression to inhibit actin polymerization, 2.5 µM Blebbistatin after completion of furrow ingression to selectively inhibit nonmuscle myosin II, and 10 µg/ml nocodazole (Sigma-Aldrich) at onset of cleavage furrow ingression.
Specimen preparation for electron microscopy
Sample preparation for electron microscopy was performed as described previously (Müller-Reichert et al., 2007). In brief, single C. elegans embryos were aspirated into cellulose capillary tubes. Capillaries of ∼1 mm in length containing early embryos were imaged under the light microscope and then placed in 100-µm-deep membrane carriers containing M9 buffer + 20% BSA (Sigma-Aldrich) as a filler. Cryoimmobilization was performed using an EM PACT2 with a rapid transfer system high-pressure freezer (Leica Microsystems). Freeze substitution was performed for ∼48–72 h at −90°C in anhydrous acetone containing 1% OsO4 and 1% uranyl acetate. Freeze substitution cocktails for drug treatment experiments contained 0.5% OsO4 and 0.5% uranyl acetate in anhydrous acetone. Specimens were then infiltrated and thin-layer embedded in epon/Araldite. Thin (90 nm) or semithick (300–350 nm) serial sections were collected on Formvar-coated copper slot grids and poststained with 2% uranyl acetate in 70% methanol followed by Reynold’s lead citrate.
Electron microscopy
Thin-section electron microscopy was performed on a 120-kV microscope (TECNAI Bio-Twin; FEI) using a 4 × 2.7–K charge-coupled device camera (Orius SC1000W; Gatan). In preparation for electron tomography, 15-nm colloidal gold particles (BBI Solutions) were attached to both surfaces of serial semithick sections to serve as fiducial markers for image alignment, and regions of interest were preselected by using a transmission electron microscope (EM 906; ZEISS) operated at 80 kV and equipped with a charge-coupled device camera (Tröndle Sharp Eye). The specimens were then placed in a tilt–rotate specimen holder (650; Gatan), and tomographic datasets were recorded using an intermediate-voltage transmission electron microscope (TECNAI F30; FEI) operated at 300 kV and equipped with a 2 × 2–K charge-coupled device camera (US1000; Gatan). The SerialEM program (Boulder Laboratory for 3D Electron Microscopy) was used to automatically acquire images every 1° over a range of ±62° at a pixel size of 1.4 nm. For dual-axis tomography, the grids were imaged in one tilt series and then rotated 90°, and a similar tilt series was acquired. Electron tomograms were computed for each tilt axis using the R-weighted back-projection algorithm (Gilbert, 1972). For double-tilt datasets, the two tomograms were aligned with each other and combined (Mastronarde, 1997). Using the IMOD software package (Boulder Laboratory for 3D Electron Microscopy), electron tomograms were displayed to generate 3D models of intercellular bridges of early embryos frozen at distinct time points throughout cytokinesis. In 3D, we modeled membranes, microtubules, microtubule ends, and vesicles. Cellular components were quantified using the program IMODinfo.
Specimen preparation and imaging by stimulated emission depletion (STED) microscopy
For STED imaging of fixed embryos, C. elegans control and tsg-101(RNAi) animals were dissected onto poly-l-lysine–coated slides, rapidly frozen in liquid nitrogen, subjected to a freeze–crack method to permeabilize the eggshell of embryos, and fixed in 100% MeOH at −20°C for 20 min as described previously (Desai et al., 2003). Antibodies directed against VPS-32 (Shen et al., 2014) and ZEN-4 (Audhya et al., 2005) were conjugated to Alexa Fluor 680 and Cy3 dyes, respectively, and incubated on the slides overnight at 4°C. Samples were mounted with ProLong Diamond and allowed to cure for 24 h at room temperature. Images were collected at room temperature using a 100× STED white oil objective (1.4 NA) on a STED 3× microscope (TCS SP8) equipped with HyD SP GaAsP detectors (Leica Microsystems) using two depletion lasers (660 and 775 nm). Image acquisition was performed using LAS X software (Leica Microsystems). Raw image files were deconvolved using Huygens Professional software (Scientific Volume Imaging), and parallel confocal images were also acquired using a white light laser.
Online supplemental material
Fig. S1 shows light and electron microscopic analysis of an embryo after treatment with nocodazole, demonstrating that microtubules are not detectable within the intercellular bridge after such treatment. Fig. S2 shows serial thin sections of tsg-101(RNAi) embryos ∼1,110 s after initiation of furrow ingression, demonstrating that ESCRT function is dispensable for the first embryonic membrane scission event in C. elegans. Fig. S3 shows light and electron microscopic analysis of an intercellular bridge after treatment with Blebbistatin, demonstrating that myosin II function is required for cytokinetic abscission in C. elegans. Videos 1, 2, 3, 4, 5, and 6 show the tomographic reconstruction and 3D modeling of the first intercellular bridge in the C. elegans embryo and correspond to Fig. 1 C. The time points of freezing after initiation of furrow ingression are Video 1, 240 s; Video 2, 375 s; Video 3, 445 s; Video 4, 650 s; Video 5, 765 s; and Video 6, 900 s.
Supplementary Material
Acknowledgments
We thank Mandy Munzig and Doreen Streichert (Faculty of Medicine Carl Gustav Carus, Technische Universität Dresden) as well as the members of the light microscopy and electron microscopy facilities at the Max Planck Institute of Molecular Cell Biology and Genetics (Dresden) for technical assistance. We are also grateful to Mihail Sarov (TransgeneOmics Unit, Max Planck Institute of Molecular Cell Biology and Genetics) for generating the TSG-101 strain.
This work was funded by Deutsche Forschungsgemeinschaft grant MU1423/4-1 to T. Müller-Reichert and National Institutes of Health grant GM088151 to A. Audhya.
The authors declare no competing financial interests.
Author contributions: All authors conceived and designed experiments. J König, E.B. Frankel, A. Audhya, and T. Müller-Reichert performed experiments and analyzed data. J. König, A. Audhya, and T. Müller-Reichert wrote the paper with input from E.B. Frankel.
Footnotes
Abbreviations used:
- ESCRT
- endosomal sorting complex required for transport
- PH
- pleckstrin homology
- STED
- stimulated emission depletion
References
- Agromayor M., and Martin-Serrano J.. 2013. Knowing when to cut and run: mechanisms that control cytokinetic abscission. Trends Cell Biol. 23:433–441. 10.1016/j.tcb.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Audhya A., Hyndman F., McLeod I.X., Maddox A.S., Yates J.R. III, Desai A., and Oegema K.. 2005. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 171:267–279. 10.1083/jcb.200506124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A., McLeod I.X., Yates J.R., and Oegema K.. 2007. MVB-12, a fourth subunit of metazoan ESCRT-I, functions in receptor downregulation. PLoS One. 2:e956 10.1371/journal.pone.0000956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballe A., and Martin-Serrano J.. 2011. ESCRT machinery and cytokinesis: the road to daughter cell separation. Traffic. 12:1318–1326. 10.1111/j.1600-0854.2011.01244.x [DOI] [PubMed] [Google Scholar]
- Carvalho A., Olson S.K., Gutierrez E., Zhang K., Noble L.B., Zanin E., Desai A., Groisman A., and Oegema K.. 2011. Acute drug treatment in the early C. elegans embryo. PLoS One. 6:e24656 10.1371/journal.pone.0024656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Tian D., Yang Y., Feng G., Cheng Z., Li W., and Ou G.. 2012. Apoptotic regulators promote cytokinetic midbody degradation in C. elegans. J. Cell Biol. 199:1047–1055. 10.1083/jcb.201209050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ L., Raiborg C., Wenzel E.M., Campsteijn C., and Stenmark H.. 2017. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem. Sci. 42:42–56. [DOI] [PubMed] [Google Scholar]
- Crowell E.F., Gaffuri A.L., Gayraud-Morel B., Tajbakhsh S., and Echard A.. 2014. Engulfment of the midbody remnant after cytokinesis in mammalian cells. J. Cell Sci. 127:3840–3851. 10.1242/jcs.154732 [DOI] [PubMed] [Google Scholar]
- Dambournet D., Machicoane M., Chesneau L., Sachse M., Rocancourt M., El Marjou A., Formstecher E., Salomon R., Goud B., and Echard A.. 2011. Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat. Cell Biol. 13:981–988. 10.1038/ncb2279 [DOI] [PubMed] [Google Scholar]
- Davies T., Jordan S.N., Chand V., Sees J.A., Laband K., Carvalho A.X., Shirasu-Hiza M., Kovar D.R., Dumont J., and Canman J.C.. 2014. High-resolution temporal analysis reveals a functional timeline for the molecular regulation of cytokinesis. Dev. Cell. 30:209–223. 10.1016/j.devcel.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., and Oegema K.. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435. 10.1101/gad.1126303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Amine N., Kechad A., Jananji S., and Hickson G.R.. 2013. Opposing actions of septins and Sticky on Anillin promote the transition from contractile to midbody ring. J. Cell Biol. 203:487–504. 10.1083/jcb.201305053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N., Fabrikant G., Kozlov M.M., and Lippincott-Schwartz J.. 2012. Computational model of cytokinetic abscission driven by ESCRT-III polymerization and remodeling. Biophys. J. 102:2309–2320. 10.1016/j.bpj.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N., Ott C., and Lippincott-Schwartz J.. 2013. Incisive imaging and computation for cellular mysteries: lessons from abscission. Cell. 155:1220–1231. 10.1016/j.cell.2013.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger A.W., Wilsch-Bräuninger M., Marzesco A.M., Bickle M., Lohmann A., Maliga Z., Karbanová J., Corbeil D., Hyman A.A., and Huttner W.B.. 2011. Proliferating versus differentiating stem and cancer cells exhibit distinct midbody-release behaviour. Nat. Commun. 2:503 10.1038/ncomms1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson S.M., and De Camilli P.. 2012. Dynamin, a membrane-remodelling GTPase. Nat. Rev. Mol. Cell Biol. 13:75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figard L., and Sokac A.M.. 2014. A membrane reservoir at the cell surface: unfolding the plasma membrane to fuel cell shape change. Bioarchitecture. 4:39–46. 10.4161/bioa.29069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger F.P., and White J.G.. 2002. Fusion and fission: membrane trafficking in animal cytokinesis. Cell. 108:727–730. 10.1016/S0092-8674(02)00668-2 [DOI] [PubMed] [Google Scholar]
- Gilbert P.F. 1972. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. II. Direct methods. Proc. R. Soc. Lond. B Biol. Sci. 182:89–102. 10.1098/rspb.1972.0068 [DOI] [PubMed] [Google Scholar]
- Glotzer M. 2005. The molecular requirements for cytokinesis. Science. 307:1735–1739. 10.1126/science.1096896 [DOI] [PubMed] [Google Scholar]
- Gould G.W. 2016. Animal cell cytokinesis: The role of dynamic changes in the plasma membrane proteome and lipidome. Semin. Cell Dev. Biol. 53:64–73. 10.1016/j.semcdb.2015.12.012 [DOI] [PubMed] [Google Scholar]
- Green R.A., Paluch E., and Oegema K.. 2012. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28:29–58. 10.1146/annurev-cellbio-101011-155718 [DOI] [PubMed] [Google Scholar]
- Green R.A., Mayers J.R., Wang S., Lewellyn L., Desai A., Audhya A., and Oegema K.. 2013. The midbody ring scaffolds the abscission machinery in the absence of midbody microtubules. J. Cell Biol. 203:505–520. 10.1083/jcb.201306036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizetti J., Schermelleh L., Mäntler J., Maar S., Poser I., Leonhardt H., Müller-Reichert T., and Gerlich D.W.. 2011. Cortical constriction during abscission involves helices of ESCRT-III–dependent filaments. Science. 331:1616–1620. 10.1126/science.1201847 [DOI] [PubMed] [Google Scholar]
- Hurley J.H. 2015. ESCRTs are everywhere. EMBO J. 34:2398–2407. 10.15252/embj.201592484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S., Wang D., and Ruvkun G.. 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 427:645–649. 10.1038/nature02302 [DOI] [PubMed] [Google Scholar]
- König J., Borrego-Pinto J., Streichert D., Munzig M., Lénárt P., and Müller-Reichert T.. 2017. Analysis of cytokinesis by electron microscopy. Methods Cell Biol. 137:225–238. 10.1016/bs.mcb.2016.03.029 [DOI] [PubMed] [Google Scholar]
- Kouranti I., Sachse M., Arouche N., Goud B., and Echard A.. 2006. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr. Biol. 16:1719–1725. 10.1016/j.cub.2006.07.020 [DOI] [PubMed] [Google Scholar]
- Lafaurie-Janvore J., Maiuri P., Wang I., Pinot M., Manneville J.B., Betz T., Balland M., and Piel M.. 2013. ESCRT-III assembly and cytokinetic abscission are induced by tension release in the intercellular bridge. Science. 339:1625–1629. 10.1126/science.1233866 [DOI] [PubMed] [Google Scholar]
- Low S.H., Li X., Miura M., Kudo N., Quiñones B., and Weimbs T.. 2003. Syntaxin 2 and endobrevin are required for the terminal step of cytokinesis in mammalian cells. Dev. Cell. 4:753–759. 10.1016/S1534-5807(03)00122-9 [DOI] [PubMed] [Google Scholar]
- Masters T.A., Pontes B., Viasnoff V., Li Y., and Gauthier N.C.. 2013. Plasma membrane tension orchestrates membrane trafficking, cytoskeletal remodeling, and biochemical signaling during phagocytosis. Proc. Natl. Acad. Sci. USA. 110:11875–11880. 10.1073/pnas.1301766110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. 1997. Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120:343–352. 10.1006/jsbi.1997.3919 [DOI] [PubMed] [Google Scholar]
- Masud Rana A.Y., Tsujioka M., Miyagishima S., Ueda M., and Yumura S.. 2013. Dynamin contributes to cytokinesis by stabilizing actin filaments in the contractile ring. Genes Cells. 18:621–635. 10.1111/gtc.12060 [DOI] [PubMed] [Google Scholar]
- McCullough J., Colf L.A., and Sundquist W.I.. 2013. Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 82:663–692. 10.1146/annurev-biochem-072909-101058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa B., and Gerlich D.W.. 2014. Cytokinetic abscission: molecular mechanisms and temporal control. Dev. Cell. 31:525–538. 10.1016/j.devcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Mishima M. 2016. Centralspindlin in Rappaport’s cleavage signaling. Semin. Cell Dev. Biol. 53:45–56. 10.1016/j.semcdb.2016.03.006 [DOI] [PubMed] [Google Scholar]
- Müller-Reichert T., Srayko M., Hyman A., O’Toole E.T., and McDonald K.. 2007. Correlative light and electron microscopy of early Caenorhabditis elegans embryos in mitosis. Methods Cell Biol. 79:101–119. 10.1016/S0091-679X(06)79004-5 [DOI] [PubMed] [Google Scholar]
- Oegema K., and Hyman A.A.. 2006. Cell division. WormBook. 10.1895/wormbook1.72.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L., O’Toole E., Schwager A., Hyman A.A., and Müller-Reichert T.. 2006. Centriole assembly in Caenorhabditis elegans. Nature. 444:619–623. 10.1038/nature05318 [DOI] [PubMed] [Google Scholar]
- Prekeris R. 2011. Actin regulation during abscission: unexpected roles of Rab35 and endocytic transport. Cell Res. 21:1283–1285. 10.1038/cr.2011.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel J.A., and Prekeris R.. 2010. Making the final cut – mechanisms mediating the abscission step of cytokinesis. Sci. World J. 10:1424–1434. 10.1100/tsw.2010.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel J.A., Park K., Morphew M.K., Reid E., Hoenger A., and Prekeris R.. 2011. Endocytic membrane fusion and buckling-induced microtubule severing mediate cell abscission. J. Cell Sci. 124:1411–1424. 10.1242/jcs.081448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel J.A., Simon G.C., Zaharris C., Weisz J., Castle D., Wu C.C., and Prekeris R.. 2012. FIP3-endosome-dependent formation of the secondary ingression mediates ESCRT-III recruitment during cytokinesis. Nat. Cell Biol. 14:1068–1078. 10.1038/ncb2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg J., Lee I.H., Iwasa J.H., and Hurley J.H.. 2017. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18:5–17. 10.1038/nrm.2016.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh A.L., and Audhya A.. 2014. The ESCRT machinery: from the plasma membrane to endosomes and back again. Crit. Rev. Biochem. Mol. Biol. 49:242–261. 10.3109/10409238.2014.881777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh A.L., Hanna M., Quinney K., Wang L., Sarkeshik A., Yates J.R. III, and Audhya A.. 2015. The VPS-20 subunit of the endosomal sorting complex ESCRT-III exhibits an open conformation in the absence of upstream activation. Biochem. J. 466:625–637. 10.1042/BJ20141202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.T., Schuh A.L., Zheng Y., Quinney K., Wang L., Hanna M., Mitchell J.C., Otegui M.S., Ahlquist P., Cui Q., and Audhya A.. 2014. Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J. Cell Biol. 206:763–777. 10.1083/jcb.201403108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman S., Kirchenbuechler D., Nachmias D., Tamir A., Werner S., Elbaum M., and Elia N.. 2016. Resolving new ultrastructural features of cytokinetic abscission with soft-X-ray cryo-tomography. Sci. Rep. 6:27629 10.1038/srep27629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D., and Pohl C.. 2014. Coupling of rotational cortical flow, asymmetric midbody positioning, and spindle rotation mediates dorsoventral axis formation in C. elegans. Dev. Cell. 28:253–267. 10.1016/j.devcel.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Thompson H.M., Skop A.R., Euteneuer U., Meyer B.J., and McNiven M.A.. 2002. The large GTPase dynamin associates with the spindle midzone and is required for cytokinesis. Curr. Biol. 12:2111–2117. 10.1016/S0960-9822(02)01390-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., and Fire A.. 1998. Specific interference by ingested dsRNA. Nature. 395:854 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Walter T., Shattuck D.W., Baldock R., Bastin M.E., Carpenter A.E., Duce S., Ellenberg J., Fraser A., Hamilton N., Pieper S., et al. 2010. Visualization of image data from cells to organisms. Nat. Methods. 7:S26–S41. 10.1038/nmeth.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Boulianne G.L., and Trimble W.S.. 2002. Membrane trafficking in cytokinesis. Semin. Cell Dev. Biol. 13:77–82. 10.1016/S1084-9521(02)00012-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.