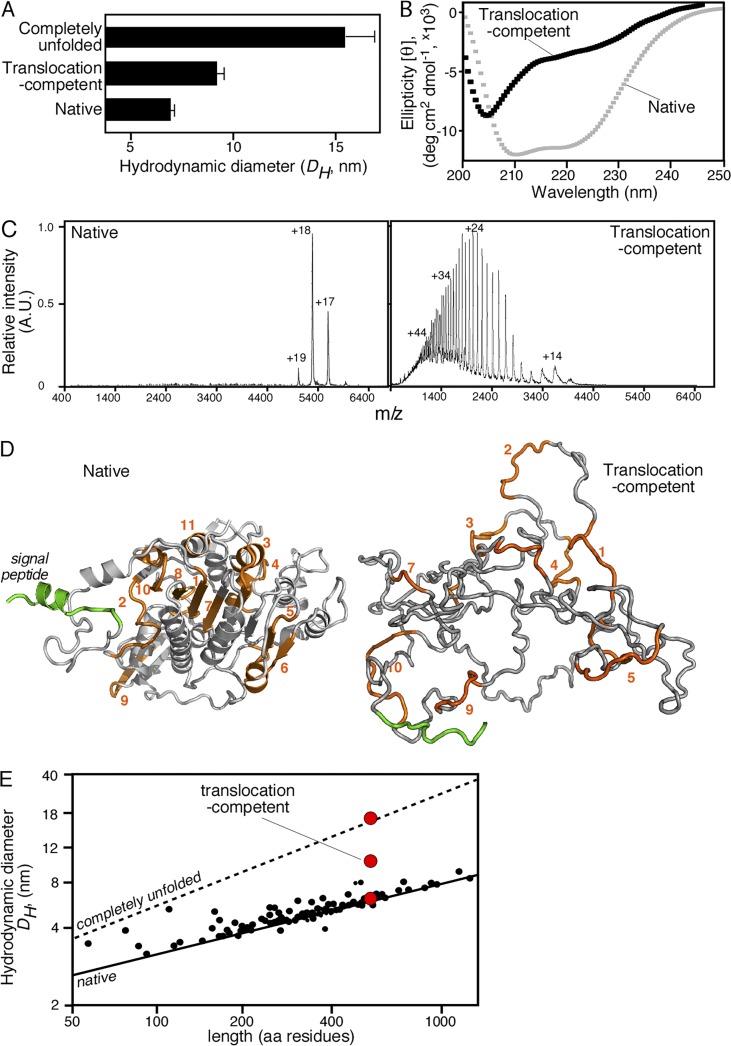
Figure 3.
Biophysical characterization of translocation-competent proPhoA. (A) Hydrodynamic diameter (DH, nanometers; x axis) of native (no urea; no DTT), translocation-competent (no urea; 1 mM DTT) and completely unfolded proPhoA (8M urea; 1 mM DTT) as determined by quasielastic laser light scattering that was performed online after gel permeation chromatography on a Superdex HR200 (see also Fig. S3 A); n = 6–15. Values represent means ± SD. For the native species, natively purified proPhoA was diluted and chromatographed in buffer L. For the translocation-competent and the completely unfolded species, urea-purified proPhoA (0.5 mM; 6 M urea), preincubated with DTT (10mM; 30min; ice), was diluted and chromatographed in buffer L supplemented with the indicated urea and DTT concentration. (B) Representative circular dichroism spectra of natively folded (no DTT) and translocation-competent (1 mM DTT) proPhoA; n > 3. x axis: wavelength (nanometers); y axis: mean residue molar ellipticity ([θ]MRW). For the translocation-competent species, urea-purified proPhoA was preincubated with 10 mM DTT (30 min; ice) and dialyzed in buffer U supplemented with 8M urea and 1 mM DTT. The natively purified proPhoA was dialyzed in 5 liters buffer U (15 h; 4°C). Spectra for both were recorded in buffer U supplemented with 1 mM EDTA, 0.2 M urea, and DTT (as indicated). Natively folded proPhoA exhibits two minima (208 and 222 nm) typical of folded, predominantly α-helical proteins, whereas the translocation-competent proPhoA does not. The urea-purified proPhoA, dialyzed in buffer U in the absence of 5 liters DTT (15 h), folds and gives spectra similar to those of the natively purified proPhoA (not depicted). (C) Representative native nano–electrospray ionization mass spectrometry spectra of native PhoA and translocation-competent proPhoA; n = 3. Translocation-competent proPhoA acquires many charges with broad distribution, typical of unfolded proteins with increased solvent-accessible surface area (Testa et al., 2013), whereas native PhoA acquires few charges with narrow distribution, typical of well-folded, compact proteins, and is a dimer. (D) Ribbon 3D model of folded E. coli PhoA (PDB: 1KHN; a single protomer is shown; left). A signal peptide (green) was modeled. Model of disordered proPhoA derived from the trigger factor–bound structure solved by nuclear magnetic resonance (Saio et al., 2014), with a DH in accordance with the quasielastic laser light scattering measurements of disordered PhoA (right). (E) Predicted and measured hydrodynamic diameters (DH) of SecA-dependent preproteins. Lines show the predicted DH of either folded (solid) or completely unfolded (dotted) preproteins as a function of their length (Wilkins et al., 1999b). Small black dots represent the calculated DH for 40 mature domains with solved structures (Table S8), using Hydropro (García De La Torre et al., 2000). Red circles represent the experimental DH measurements for proPhoA (as indicated; see also A and Fig. S3 B).