Figure 4.
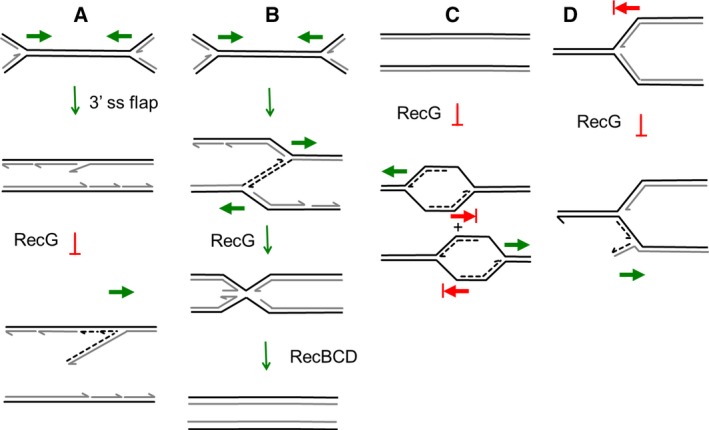
Four different models proposed to explain how RecG controls DNA amplification. (A) Fork collision and restart at a 3′ flap. When two replication forks (moving in the directions of the green arrows) collide, it is hypothesised that in the absence of RecG a 3′ flap is generated that leads to the assembly of a replication fork. In the presence of RecG, the 3′ flap is converted into a 5′ flap that can be degraded by 5′–3′ exonucleases 18, 19, 21, 22. (B) Fork collision and template‐switching followed by replication fork reversal. When two replication forks (moving in the directions of the green arrows) collide, it is hypothesised that template switching occurs leading to over‐replication. This is corrected by RecG‐dependent replication fork reversal and DNA degradation at one (or both) of the replication forks 23. (C) cSDR and termination at Tus/ter blocks. It is proposed that, in the absence of RecG, cSDR initiates at sites of transcription around the genome leading to replication forks that are blocked by Tus/ter. This results principally in over‐replication of the region between termination sites (at the positions of blocked red arrows) as cSDR forks are removed by colliding with origin‐initiated replication forks 25. (D) Reverse‐restart of an arrested replication fork. At an arrested replication fork (at the position of the blocked red arrow) RecG prevents the assembly of the replicative helicase on the newly synthesised lagging‐strand. In the absence of RecG, this loading is permitted and backwards‐directed DNA replication occurs 17.
