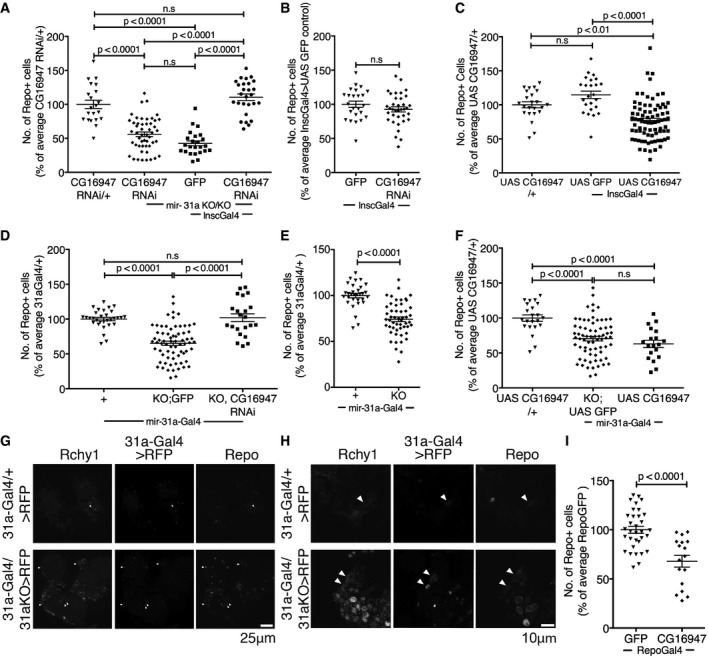
Number of anti‐repo‐positive glia in adult brains at 7 days. Glia counts in the experimental samples are represented as a percentage of the average number of glia in central brains of the indicated controls. Data were analysed using one‐way ANOVA with
post hoc Tukey analysis (A, C, D, F) or with an unpaired Student's
t‐test (B, E). Error bars represent SEM. (A) The
UAS‐CG16947 RNAi transgene without a Gal4 driver was used as a control.
miR‐31a KO/KO indicates the homozygous deletion mutant. A
UAS‐GFP transgene was used as a control for expression of the RNAi transgene with
Insc‐Gal4 in the mutant background. ns: not significant. See also
Appendix Fig S1 and
Table EV1. (B) Expression of
UAS‐GFP or
UAS‐CG16947 RNAi using
Insc‐Gal4 cells in an otherwise normal background. (C) The
UAS‐CG16947 transgene without a Gal4 driver was used as a control.
UAS‐GFP was used as a control for expression of the
UAS‐CG16947 transgene with
Insc‐Gal4 in an otherwise normal background. (D) All samples carried one copy of the
miR‐31a‐Gal4 allele. KO;
GFP indicates the deletion allele and a
UAS‐GFP transgene. KO,
CG16947 RNAi indicates the deletion allele and the
UAS‐RNAi transgene to deplete
CG16947 mRNA. (E) All flies carried the
miR‐31a‐Gal4 allele. KO indicates the deletion allele. (F) The
UAS‐CG16947 transgene without a Gal4 driver was used as a control.
UAS‐GFP in the mutant background was used for comparison to expression of
UAS‐CG16947 transgene with
miR‐31a‐Gal4 in an otherwise normal background.