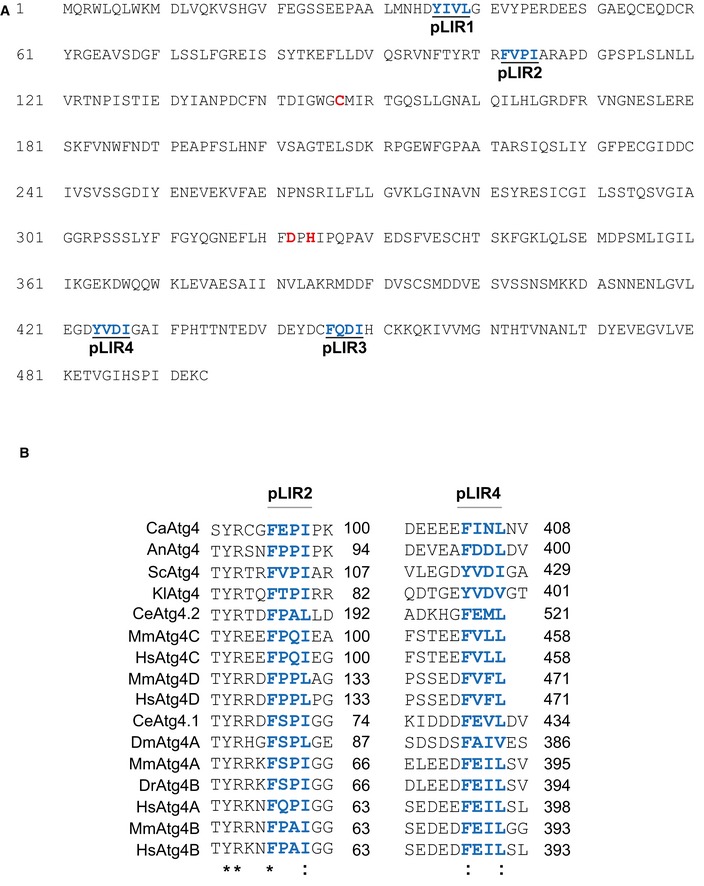
The amino acid sequence of the regions flanking the two conserved pLIR motifs of
S. cerevisiae Atg4 (in blue), that is, F102 to I105 (pLIR2) and Y424 to I427 (pLIR4), were aligned with that of homologous proteins from different species using the Kalign alignment tool (
http://www.ebi.ac.uk/Tools/msa/kalign/). UniprotKB accession numbers are
C. albicans Atg4 (
Q59UG3),
A. nidulans Atg4 (
Q5B7L0),
S. cerevisiae Atg4 (
P53867),
K. lactis Atg4 (
Q6CQ60),
C. elegans Atg4.1 (
Q9NA30),
C. elegans Atg4.2 (
Q9U1N6),
D. melanogaster Atg4 (M9PBM3),
D. rerio Atg4B (
Q6DG88),
M. musculus ATG4A (
Q8C9S8),
M. musculus ATG4B (
Q8BGE6),
M. musculus ATG4C (
Q811C2),
M. musculus ATG4D (
Q8BGV9),
H. sapiens ATG4A (
Q8WYN0),
H. sapiens ATG4B (
Q9Y4P1),
H. sapiens ATG4C (
Q96DT6), and
H. sapiens ATG4D (
Q86TL0). The asterisk indicates conservation of the residue while two dots designate similarity.