Figure 8. Models for the structure of yeast Atg4 catalytic site and for Atg4 deconjugation activity on autophagosomal membranes.
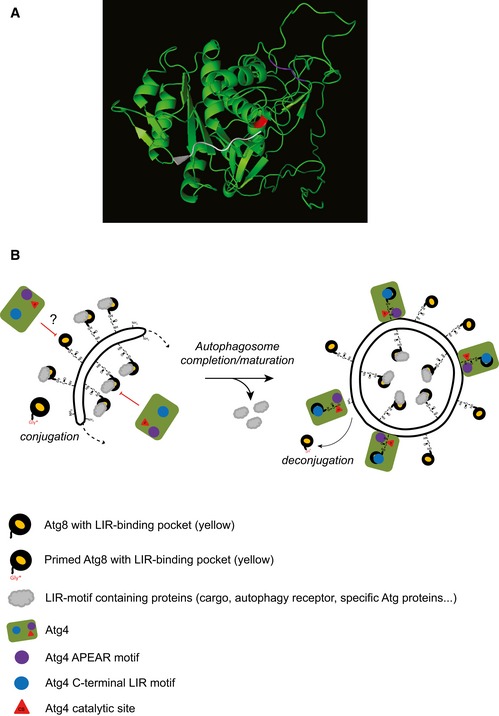
- Three‐dimensional model predicting yeast Atg4 structure generated using the RaptorX online program (http://raptorx.uchicago.edu/StructurePrediction/predict/) 55, which covered Atg4 sequence from aa 1–403 leaving C‐terminal region with 91 aa unpredicted. The catalytic site (C147) of Atg4 is highlighted in red while APEAR is colored in purple. The last eight C‐terminal amino acids of LC3B, in white, were positioned into the catalytic site as shown for ATG4B 37.
- After proteolytic priming, the C‐terminal glycine of Atg8 gets activated via the ubiquitin‐like conjugation system and linked through an amide bond to the PE present on autophagosomal membranes. Atg8‐PE is involved in autophagosome biogenesis at the PAS (dotted arrow). At this location, Atg8‐PE also associates with various proteins (gray clouds), such as cargo receptors, by binding their LIR motifs through a defined structural pocket (yellow). Atg4 binding to Atg8‐PE has to be controlled to avoid premature deconjugation. Occupation of the LIR motif‐binding pocket by other factors and/or other regulatory mechanisms such as post‐translational modifications (depicted with a question mark) could inhibit Atg4 action on autophagosomal membranes. As soon as the autophagosome is completed, the Atg machinery, including the shielding factors, is released allowing Atg4 access to autophagosomal membranes. This latter event involves both Atg4 binding to Atg8 via cLIR (blue circle) and association of the APEAR motif (purple circle) possibly with the C‐terminal region of Atg8‐PE, which allows the correct positioning of the C‐terminus into the catalytic site (red triangle) of Atg4.
