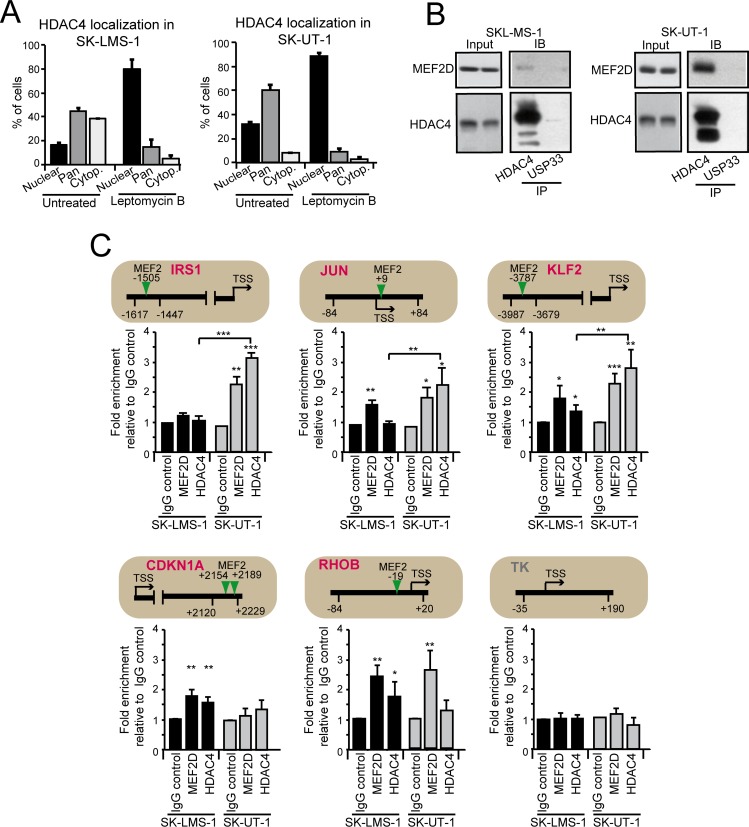
Fig 4. Analysis of MEF2D-HDAC4 repressive complexes in LMS cells.
A) Quantitative analysis of the immunofluorescence studies. LMS cells were treated or not for 2 hours with 5ng/ml leptomycin B (LC Laboratories). After fixation of the cells, immunofluorescence analysis was performed to visualize HDAC4. Nuclei were stained with Hoechst 33342. Data are presented as mean ± SD (n = 3). B) MEF2D-HDAC4 complexes were immunoprecipitated using 1μg of anti-HDAC4, or anti-USP33, as a control, antibodies. Immunoblotting using an anti-MEF2D antibody was next used for the detection. The same amounts of cellular lysates were immunoprecipitated and the immunoblot were developed under the same circumstances. C) Chromatin was immunoprecipitated from SK-LMS-1 or SK-UT-1 cells using the anti-MEF2D and the anti-HDAC4 antibodies. Anti-FLAG antibody was used as control. TK promoter was used as negative control. The MEF2 binding site, the amplified region and the TSS are indicated for each tested gene, respectively with a vertical arrow, two arrowheads and a horizontal arrow. The TK promoter was used as negative control.