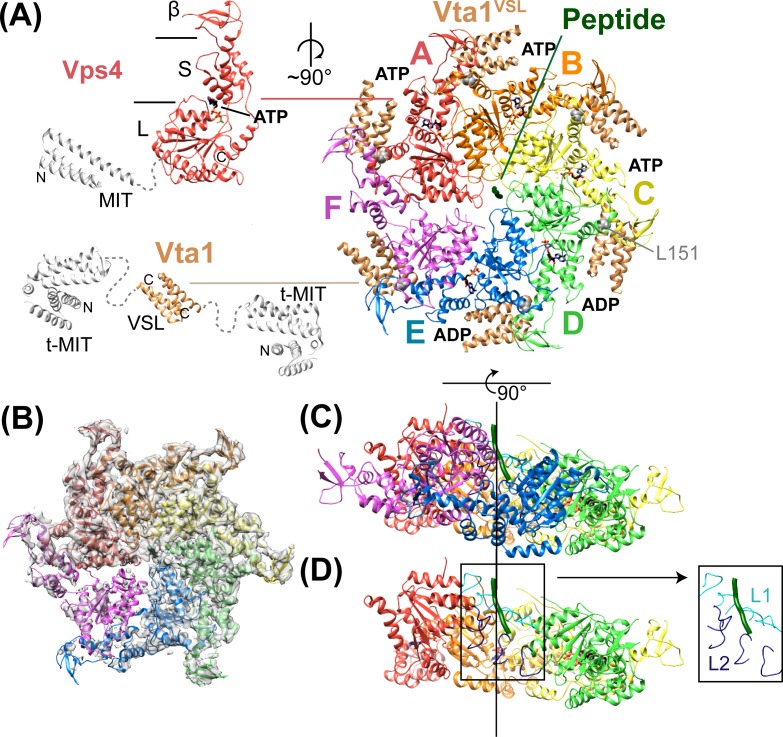
Figure 2. Structure of Vps4101-437:Vta1VSL:ESCRT-IIIpeptide:ADP·BeFx.
(A) Structure of the complex. The Vps4 and Vta1 constructs used for cryo-EM structure determination are shown in color on the left, with excluded segments colored white. MIT, large AAA ATPase (L), small AAA ATPase (S) and β domains of Vps4 are labeled, as are the t-MIT and VSL domains of the Vta1 dimer. L151, a residue critical for hexamerization, is shown in gray spheres. (B) 4.3 Å map with the Vps4 model. (C) Side view of Vps4 hexamer, oriented with the subunit A-D helix axis vertical (black line). (D) Same as panel D but with subunits E and F removed. The inset shows the position of pore loops 1 (L1, residues 203–210, cyan) and pore loops 2 (L2, 240–248, dark blue) relative to the ESCRT-III peptide (dark green).