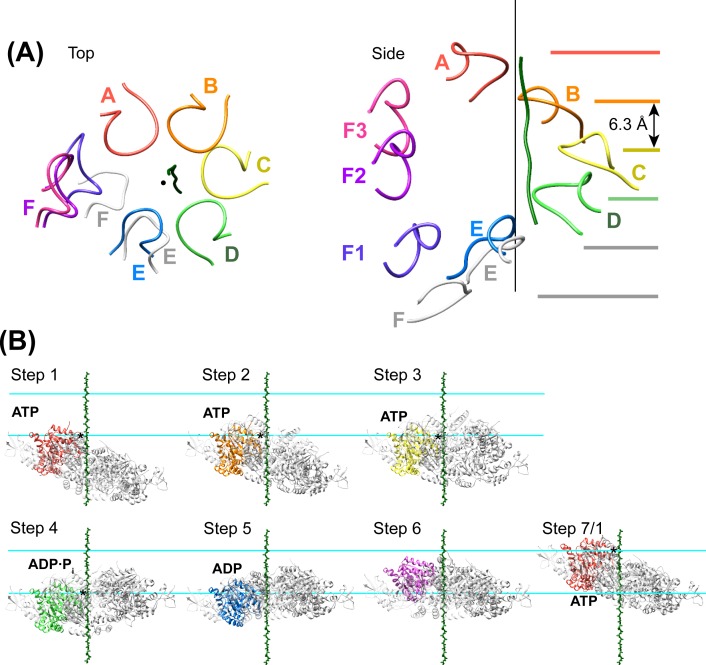
Figure 7. Peptide binding and mechanism of translocation.
(A) The pore loop 1 residues of subunits A-D form a helix (axis, black line) that matches the symmetry of a canonical twisted β-strand, which rotates 60° and translates 6.3 Å every two residues. In white are the positions that subunits E and F would adopt if they continued this helix. The three positions seen for subunit F (Figure 2—figure supplements 7–8) appear to be snapshots along the return path from the end of the helix at subunit E to the start of the helix at subunit A. (B) Steps along the translocation cycle inferred from the cryo-EM structure. The peptide shown is modeled as a β-strand along the helix axis of subunits A-D. Vps4 maintains a constant interaction with the peptide through steps 1 to 4 before dissociating at step 5 and rebinding 12 residues further up the peptide at step 7, which is equivalent to step 1. Nucleotides suggested by density and coordination geometry are labeled. Pore loop 1 contacts with the substrate peptide in steps 1–4 are indicated with an asterisk. The two subunits closest to the view direction are included with 50% transparency. The two horizontal lines are separated by 37.8 Å (12 residues) and indicate points of substrate contact with pore loop 1 of the highlighted subunit.