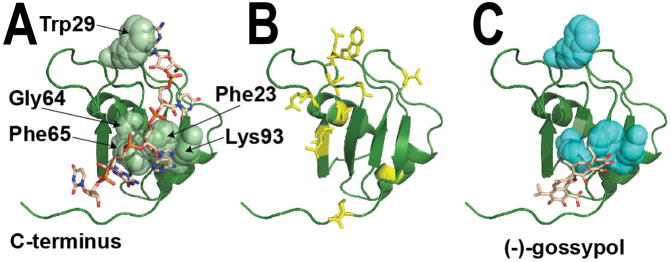
Figure 3. The NMR structure of RRM1 from mouse Msi1.
(A) The structure of this RRM domain (indicated as a green ribbon) was determined in complex with a short segment of cognate RNA (sequence GUAGU, indicated as brown sticks), PDB ID 2rs2 (62). The RRM domain is a conserved fold that packs two α-helices against one face of a four-stranded β-sheet. As with other “canonical” RRMs (88), the opposite face of this β-sheet is then used to bind a single-stranded segment of RNA, as shown. The residues with the largest differences in NMR HSQC peak intensities upon addition of RNA include Phe23, Trp29, Gly64, Phe65, and Lys93 (shown in spheres), confirming the role of these residues in Msi1 binding to RNA (76,78). Structures of other proteins containing tandem RRM domains often show these domains in an arrangement that presents a single deep RNA-binding cleft (88,89). In this Msi1-RRM1 structure, the C-terminus of the RRM1 domain makes close contact with the RNA beyond simply the canonical interactions on the β-sheet. Thus, the structural basis for Musashi-RNA association may also be strongly affected by interactions between the two RRM domains that are not captured in the complex shown here. (B) Upon addition of oleic acid to the Musashi RRM domain, the NMR HSQC peak intensity for residues throughout the structure exhibit diminished intensity (cyan sticks), suggesting extensive structural changes consistent with allosteric inhibition (76). (C) Upon addition of (−)-gossypol to the RRM domain, the NMR HSQC peak intensity is diminished primarily for residues involved in RNA binding (cyan spheres), suggesting direct inhibition and supporting the docked model shown (78).