Figure 1.
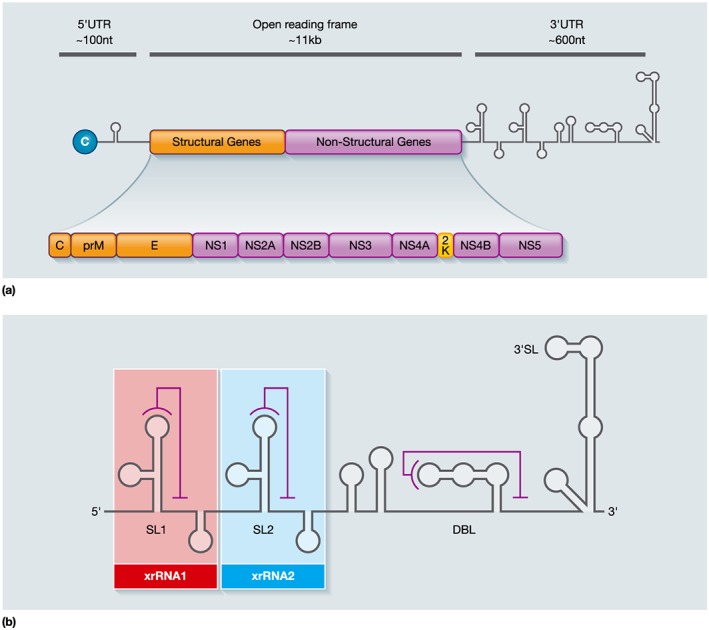
(a) Organisation of the flavivirus genome. The flavivirus genome is composed of a single‐stranded, positive‐sense RNA, of approximately 11 kb. The single open reading frame contains the three structural proteins (C: capsid, prM: premembrane, E: envelope) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). These are flanked on either side by highly structured 5′ and 3′ untranslated regions. The gene products are generated from the single polyprotein by co‐ and posttranslational cleavage. This also results in the production of the 2K peptide between NS4A and NS4B. (b) Structure of ZIKV subgenomic flavivirus RNA (sfRNA), as predicted following structural studies and RNA folding analysis. Although the structure of sfRNA varies for different flaviviruses, they all contain similar motifs. All flavivirus sfRNAs contain stem loop (SL) and dumbbell (DBL) structures, which consist of conserved nucleotides capable of forming pseudoknots (PK). PK are represented by lines. Two sfRNAs of differing size are produced during ZIKV infection due to the stalling of XRN1 at the SL structures. Predicted sfRNAs: stalling at SL1 produces xrRNA1 (red box), and xrRNA2 (blue box) is produced by stalling at SL2 (Akiyama et al., 2016; Donald et al., 2016)
