Figure 1.
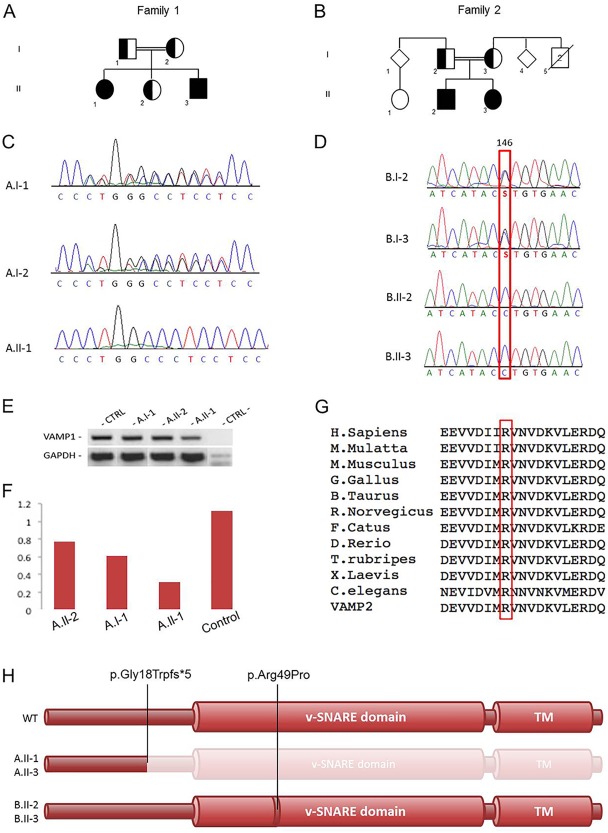
Family trees, Sanger sequencing, and VAMP1 mutation analysis. (A) Pedigree from Family 1. (B) Pedigree from Family 2. (C) Electropherograms of carrier parents and index case with the c.51_64delAGGTGGGGGTCCCC variant. (D) Electropherograms of carrier parents and the 2 patients with the c.146G>C variant. (E) Reverse transcription polymerase chain reaction (PCR) amplifying the mutant cDNA transcript from mRNA extracted from the immortalized lymphoblastoid cell lines of the index case, her father, and her healthy sister (both carriers of the heterozygous deletion), and a wild‐type control (CTRL). (F) Analysis of the semiquantitative PCR using the densitometry software ImageJ after normalization relative to a housekeeping gene (GAPDH) and calculation using a relative relationship method. (G) Multiple‐sequence alignment showing complete conservation of protein sequence across species and SNARE homolog VAMP2 in the v‐SNARE coiled coil homology, in which the disease‐segregating mutation p.Arg49Pro was found. (H) VAMP1 protein representative. The c.51_64delAGGTGGGGGTCCCC deletion causes a nonsense mutation, putatively producing a truncated protein lacking the v‐SNARE and the transmembrane (TM) domains, whereas the p.Arg49Pro mutation affects an active site of the conserved v‐SNARE domain.
