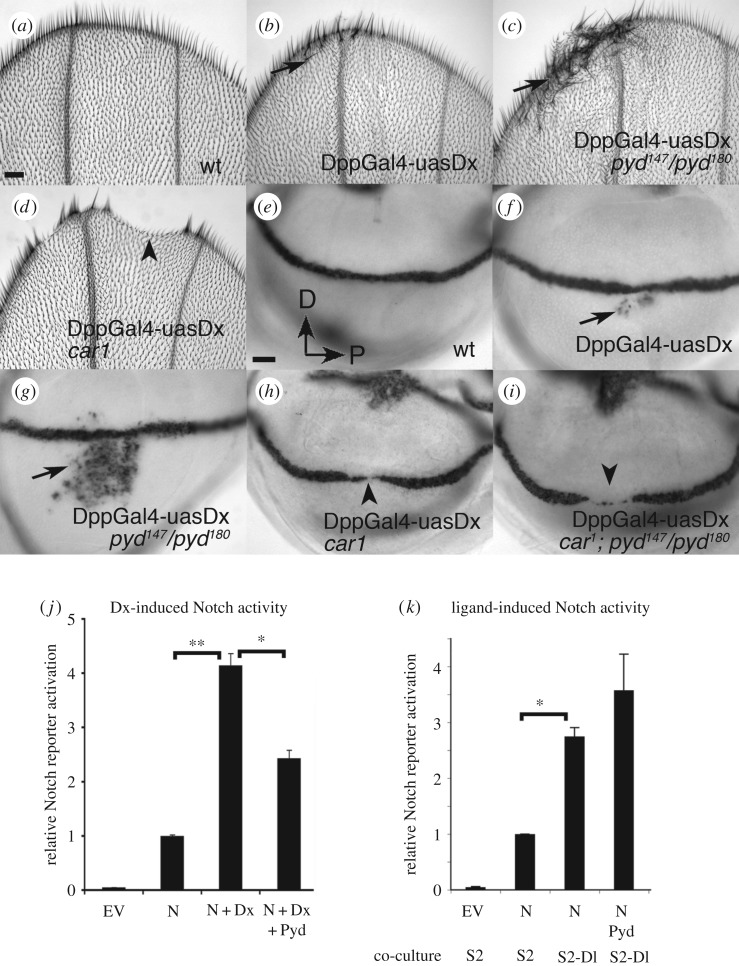
Figure 1.
Pyd suppresses Notch signalling induced by Dx expression. Adult distal wing tips showing a wild-type (wt) margin (a), and ectopic margin bristles (arrows) induced by overexpression of Dx with Dpp-Gal4, in a wt background (b), or pyd147/pyd 180 null (c). (d) shows notching associated with overexpression of Dx in a car1 mutant (arrowhead). (e–i) Late third instar wing imaginal discs stained for wingless mRNA expression to report Notch activation. In a wt disc (e) there is a clear line of wingless expression demarcating the dorsal/ventral boundary. (f–i) Dx overexpressed along the A/P axis using the Dpp-Gal4 driver. (f) Mild ectopic wingless expression (arrow) induced in wt background. (g) Enhanced wingless expression in pyd null mutant. Overexpression of Dx in a car1 mutant (h) or in a car1, pyd double mutant (i), leads to a loss of wingless (arrowheads). Scale bar in (a) represents 40 µm in (a–d). Scale bar in (e) represents 20 µm in (e–i). (j) Pyd suppresses Dx-induced signalling in S2 cells. S2 cells were transfected with empty vector (EV) or as indicated. Normalized mean Notch reporter activity measured by luciferase reporter expression is relative to cells expressing Notch only. Error bars are s.e.m., n = 3, * or ** indicates p < 0.05 by Student t-test for comparisons as indicated. (k) Pyd does not suppress ligand-induced signalling. Luciferase assay in S2 cells showing the consequence of Pyd expression on ligand-induced Notch activation. Normalized mean Notch reporter activity is shown relative to cells expressing Notch co-cultured with non-expressing S2 cells. S2 cells were transfected as indicated. Data represent mean of five experimental repeats, each performed in triplicate. Error bars are s.e.m., * indicates p < 0.05, Student t-test for comparison as indicated on graph.