Abstract
Ubiquitination is a versatile post-translational modification that regulates a multitude of cellular processes. Its versatility is based on the ability of ubiquitin to form multiple types of polyubiquitin chains, which are recognized by specific ubiquitin receptors to induce the required cellular response. Linear ubiquitin chains are linked through Met 1 and have been established as important players of inflammatory signalling and apoptotic cell death. These chains are generated by a ubiquitin E3 ligase complex called the linear ubiquitin chain assembly complex (LUBAC) that is thus far the only E3 ligase capable of forming linear ubiquitin chains. The complex consists of three subunits, HOIP, HOIL-1L and SHARPIN, each of which have specific roles in the observed biological functions of LUBAC. Furthermore, LUBAC has been found to be associated with OTULIN and CYLD, deubiquitinases that disassemble linear chains and counterbalance the E3 ligase activity of LUBAC. Gene mutations in HOIP, HOIL-1L and OTULIN are found in human patients who suffer from autoimmune diseases, and HOIL-1L mutations are also found in myopathy patients. In this paper, we discuss the mechanisms of linear ubiquitin chain generation and disassembly by their respective enzymes and review our current understanding of their biological functions and association with human diseases.
Keywords: ubiquitin, RBR E3 ligases, LUBAC, OTULIN, immune responses, inflammatory signalling
1. Introduction
Post-translational modifications of proteins extend their functional landscape and allow rapid changes in their behaviour in response to stimuli without the need for protein synthesis de novo. Ubiquitination is one of the most diverse forms of post-translational modification. It plays crucial roles in nearly every type of cellular function including protein degradation, DNA damage responses, trafficking and intracellular signalling. Its diversity springs from its ability to modify target proteins either with a single or multiple single ubiquitin molecules (mono- or multi-monoubiquitination) or with polyubiquitin chains, in which individual ubiquitin molecules are linked to one another via their C-terminal carboxyl group and either a lysine side chain or the N-terminal amino group of methionine 1 (Met 1). In total, eight different types of homotypic polyubiquitin chains are formed plus many different combinations of mixed and branched ubiquitin chains [1]. The last type of homotypic polyubiquitin chains identified were the linear, also called Met 1-linked, chains in which a peptide bond connects the ubiquitin molecules within the oligomer. In 2006, a multi-subunit complex termed linear ubiquitin chain assembly complex (LUBAC) was identified to be responsible for their synthesis [2] and it was later shown that they are required for the activation of the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) transcription factor and play important roles in immune and inflammatory signalling processes [3,4]. Since then, their functional roles have been extended to include regulation of cell death, T- and B-cell development, mouse embryonic development, heat tolerance in flies, and cancer and autoimmune diseases in humans [5–10].
Modification of proteins with ubiquitin requires the activity of three different enzymes that act in a sequential cascade that includes E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases (figure 1a) [11]. E3 ligases are the key determinants of the ubiquitination process and confer specificity as they select the target protein and in some cases also the type of ubiquitin modification [12–14]. This process is reversible and ubiquitin can be removed from a target by deubiquitinating enzymes (DUBs) [15,16]. Often polyubiquitin chain synthesis and cleavage occur in unison, thereby ensuring that the correct type of ubiquitin modification is available at the required time and place.
Figure 1.
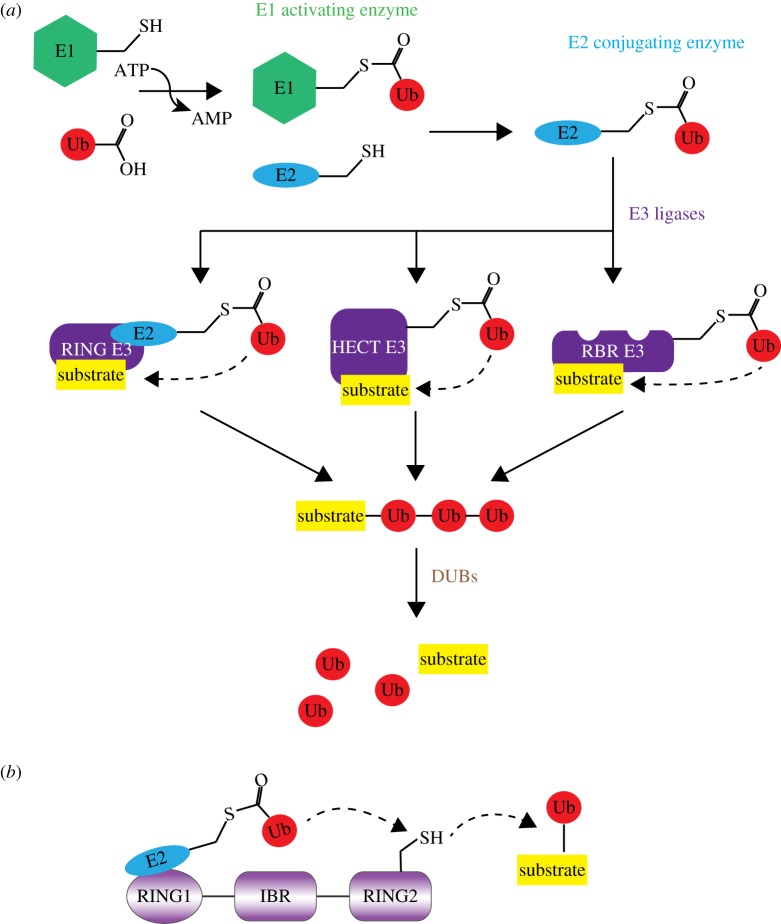
The ubiquitination cascade. (a) Schematic depiction of the ubiquitin conjugation system showing the ATP-dependent activation of ubiquitin and thioester formation with E1 and its subsequent transfer onto E2. Transfer onto the substrate is catalysed by E3 ubiquitin ligases, which are divided into three classes: RING, HECT and RBR ligases. Ubiquitination is reversible and ubiquitin chains are removed from substrates by DUBs. (b) Domain organization of the RBR catalytic module showing the RING1, IBR and RING2 domains. The E2∼Ub conjugate is recognized by the RING1 domain and transferred to a conserved cysteine in RING2 to form a thioester intermediate before the final transfer of ubiquitin onto the substrate.
E3 ubiquitin ligases catalyse the transfer of ubiquitin onto the substrate using different mechanisms and, based on this property, have been classified into three different subfamilies (figure 1a). Really interesting new gene (RING)-type ligases play a more indirect role and bring the substrate and ubiquitin-loaded E2 (E2∼Ub) together, and ubiquitin transfer occurs directly from the E2 onto the substrate [12–14]. By contrast, the homologous to the E6AP carboxyl terminus (HECT)-type ligases take the ubiquitin from the E2 to form a thioester intermediate before its final transfer onto the substrate [13,14,17]. E3 ligases of this family determine the topology of the polyubiquitin chain formed, independent of the cognate E2 [17]. A third class of E3 ligases combines properties of both these types and employs a RING domain (RING1) to initially recognize the ubiquitin-loaded E2, which is subsequently transferred to a conserved active-site cysteine in a RING2 domain to form a thioester intermediate, before the final, E2-independent ubiquitin transfer onto the substrate. This mechanism is adopted by the RING-between-RING (RBR) family of E3 ligases (figure 1b) [18–20]. Unlike other types of homotypic ubiquitin chains that can be synthesized by multiple HECT E3s and E2-RING E3 combinations, linear ubiquitin chains are only produced by the multi-subunit ligase LUBAC, which is a member of the RBR family of E3s.
The last few years have seen a big increase in our understanding of mechanistic and structural features of LUBAC activity and the physiological role of linear ubiquitin chains in different organisms. In this review, we will focus on these recent advances and discuss our current understanding of the determinants and regulators of linear chain synthesis by LUBAC and cleavage by DUBs and their role in the regulation of a multitude of cellular processes.
2. Composition of LUBAC
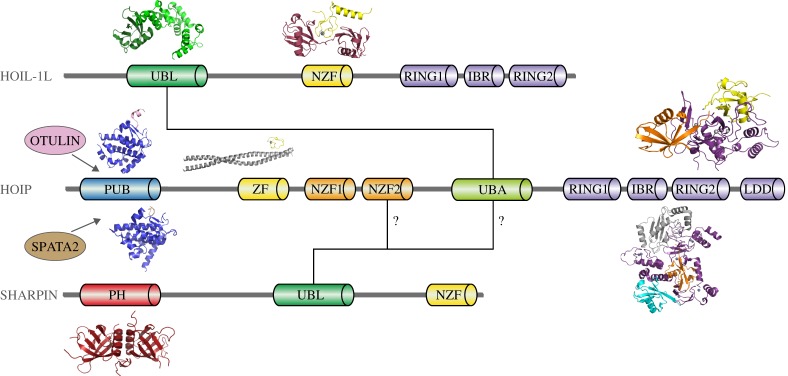
When LUBAC was discovered in 2006, it was initially thought to be composed of only two subunits termed HOIL1-interacting protein (HOIP)/RNF31 and haem-oxidized IRP2 ubiquitin ligase1L (HOIL-1L)/RBCK1, which associate into a high-molecular-weight complex of unknown stoichiometry [2]. Since then, work by multiple groups has revealed that the composition of LUBAC is more elaborate and contains an additional non-catalytic subunit called Shank-associated RH domain-interacting protein (SHARPIN) [21–23]. Moreover, the HOIP subunit of LUBAC has been shown to associate with regulatory proteins including the linear chain-specific OTU domain DUB with linear linkage specificity (OTULIN)/Gumby/FAM105B and the adaptor protein spermatogenesis-associated protein 2 (SPATA2), which links LUBAC to another DUB, CYLD, which cleaves linear and Lys 63-linked polyubiquitin chains [24–27]. This multi-protein machinery is held together by defined protein–protein interactions as illustrated in figure 2, some of which have been characterized structurally [27–30]. Nevertheless, it is currently unknown if any of the individual constituents of LUBAC are present in multiple copies and if the stoichiometry of the complex may be important for its physiological function. Furthermore, HOIP, HOIL-1L and SHARPIN contain ubiquitin-binding domains (UBDs) of varying specificity [4,21,31]. Nevertheless, how binding to different types of polyubiquitin chains contributes to LUBAC stability, localization and/or regulation of catalytic activity is still unknown.
Figure 2.
Composition of LUBAC. Schematic of the domain composition of HOIP, HOIL-1L and SHARPIN. The domains mediating the interaction between subunits are highlighted. The UBL domain of HOIL-1L binds the UBA domain of HOIP, while the interaction between SHARPIN and HOIP is less well defined and may include the NZF2 or UBA domains of HOIP, as indicated by question marks. Structural information exists for a number of subdomains of LUBAC components and their complexes with binding partners. Structures are shown where available and include the HOIL-1L UBL/HOIP UBA complex (4DBG), the HOIL-1L NZF/linear diUb complex (3B08), HOIP PUB/OTULIN PIM (4OYK, 4P0W), HOIP PUB/SPATA2 PIM (5LJN), HOIP ZF/NEMO UBAN (4OWF), HOIP RING2-LDD/ubiquitin (4LJO), HOIP RBR/UBE2D2-ubiquitin (5EDV) and the PH domain of SHARPIN (4EMO).
Interaction between the HOIP and HOIL-1L subunits is mediated by their respective ubiquitin-associated (UBA) and ubiquitin-like (UBL) domains, which form a non-canonical UBA–UBL complex as revealed in their crystal structure (figure 2) [28]. The UBL domain of HOIL-1L adopts the typical ubiquitin fold, whereas the UBA of HOIP forms a nine-helix bundle that includes a three-helix bundle consisting of α6, α7 and α8 that is similar to canonical UBA domains. Intriguingly, in the crystal HOIP UBA makes contacts with three different molecules of HOIL-1L UBL that all recognize different binding surfaces on HOIP. Only one of these involves the Ile 44 hydrophobic patch that is normally involved in UBL-mediated interactions. However, binding studies combined with deletion mapping showed that complex formation occurs with a 1 : 1 stoichiometry in solution and involves helices α6, α7, α8 and α9, which recognize a surface on the UBL that is opposite the canonical UBA-binding surface. SHARPIN also employs its UBL domain to bind HOIP. However, the region of HOIP mediating this interaction is unclear and both UBA and NZF2 have been suggested to be involved [21–23].
The N-terminal portion of HOIP contains a PUB domain, which mediates the interaction with the PUB interaction motifs (PIMs) of OTULIN and SPATA2 (figure 2). Crystal structures of the HOIP PUB domain bound to the PIMs of OTULIN and SPATA2 have revealed that they both recognize the same binding site in HOIP and hence that their interaction is mutually exclusive [27,29,30]. PIMs contain a conserved tyrosine and its phosphorylation has been shown to abrogate the interaction with OTULIN in vitro, suggesting a mechanism for how the interaction might be regulated. However, a physiological relevance of phosphorylation has not been shown so far nor has a kinase capable of phosphorylating OTULIN been identified. The recent discovery of SPATA2 as a binding partner of HOIP solved the conundrum that CYLD was known to play a functional role in LUBAC signalling and required the PIM-binding capability of the PUB domain of HOIP [32,33] but that no direct interaction between the two proteins could be detected in vitro. Instead, it has now been shown that SPATA2 itself contains a PUB domain that unlike other PUBs does not recognize PIMs but instead interacts with CYLD to connect this DUB to LUBAC [26,27]. The biological role of these interactions between LUBAC components and DUBs are described in detail in §6.
3. The linear ubiquitin chain synthesis machinery
The HOIP subunit of LUBAC contains all the catalytic machinery required to synthesize linear ubiquitin chains with high specificity [2,3]. This activity is located in its C-terminal portion within the RBR domain plus a C-terminal extension referred to as the linear ubiquitin chain-determining domain (LDD) that is specific to HOIP [34,35]. RBR domains have a conserved domain structure that consists of an N-terminal RING1 domain that recognizes the ubiquitin-loaded E2, a central in-between RING (IBR) domain of yet unknown function and a C-terminal RING2 domain that harbours the catalytic cysteine, which forms the thioester intermediate during ubiquitin transfer (figure 1b) [18]. Recent structural and biochemical studies have allowed first insight into individual steps of the ubiquitin transfer reaction from E2 to E3 (HOIP) and onto a ubiquitin substrate, and have provided a mechanistic explanation for the in vitro and in vivo observed chain linkage specificity [34–37].
To modify a target with a polyubiquitin chain, monoubiquitination must occur first to initiate a chain, which subsequently can be extended. Originally, it was suggested that HOIP, possibly in conjunction with another LUBAC subunit, directly recognizes the target and attaches the first ubiquitin for chain initiation after which ubiquitin becomes the substrate during chain extension [3,38,39]. More recently, however, an alternative mechanism was proposed, in which the protein target, such as the IκB kinase (IKK) subunit NEMO, is modified by a Lys 63-linked chain first (by an as yet unidentified E3), which is then extended by LUBAC with a linear chain [40]. In such a model, the only substrate of LUBAC is ubiquitin itself. At present, it is not clear if these models are mutually exclusive or if a given stimulus determines if homotypic linear or mixed chains are produced.
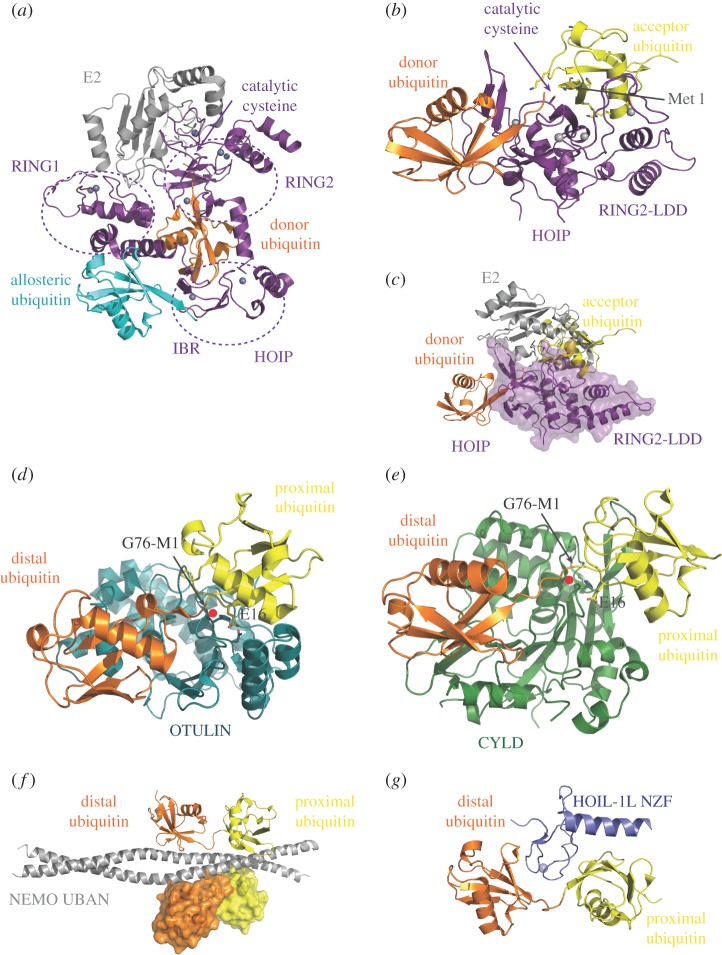
The mechanistic features that determine the highly specific synthesis of linear ubiquitin chains have been uncovered in the high-resolution crystal structure of the RING2-LDD fragment of HOIP in complex with ubiquitin [36]. This structure contains both the donor and acceptor ubiquitins and provides a snapshot of the last step of the ubiquitination process in which ubiquitin is transferred from the E3 thioester onto the growing ubiquitin chain. The acceptor ubiquitin is accommodated by a surface created by the RING2-LDD fragment and oriented such that its N-terminal amino group is in very close proximity to the active-site cysteine (Cys 885, 3.5 Å) (figure 3b). In this arrangement, none of the seven lysine residues of ubiquitin is sufficiently close to Cys 885 to compete with the α-amino of Met 1 for a nucleophilic attack on the thioester intermediate, explaining the observed high linear chain specificity. Furthermore, the structure revealed that the LDD is not an isolated domain that confers specificity for linear ubiquitin chain synthesis. Instead the LDD consists of a ZF (ZF1), which is integrated into RING2 of HOIP with a helical extension (the ‘helical base’), both of which are required to correctly position the acceptor ubiquitin. This arrangement indicates that the LDD could not simply be transferred onto another E3 to bestow linear chain specificity and explains why LUBAC is the only E3 capable of forming linear ubiquitin chains. A histidine residue in the active site, close to the catalytic cysteine, has been shown to be essential for ubiquitin transfer, though not for formation of the HOIP∼ubiquitin thioester intermediate, suggesting that it acts as a general base to deprotonate the incoming nucleophile (the α-amino of Met 1) [36]. A histidine in this position is conserved in a number of RBR domains where it probably plays the same role.
Figure 3.
Structures of protein complexes mediating linear chain formation, disassembly and recognition. (a) Structure of the HOIP RBR/UBE2D2-ubiquitin complex in its monomeric form with the RBR domain shown in purple with its subdomains indicated the E2-conjugating enzyme in grey, the conjugated ubiquitin in orange and the allosteric ubiquitin in cyan (5EDV). (b) Structure of the active HOIP RING2-LDD/ubiquitin complex showing the position of donor (orange) and acceptor (yellow) ubiquitin. The positions of the catalytic cysteine and Met 1 of the acceptor ubiquitin are indicated (4LJO). (c) Clash between the binding sites of the HOIP RING2-LDD fragment for acceptor ubiquitin and E2, indicating that the E2 has to dissociate before the ubiquitin can be transferred onto the growing chain. (d,e) Structures of the OTU domain of OTULIN (d, 3ZNZ) and USP domain of CYLD (green) (e, 4WXF) bound to linear diubiquitin with the distal ubiquitin in orange and the proximal ubiquitin shown in yellow. The positions of the G76-M1 bond to be cleaved, the catalytic cysteine (red dot) and Glu 16 from the proximal ubiquitin are indicated. (f) Structure of the UBAN domain of NEMO (grey) bound to linear diubiquitin with the distal and proximal ubiquitin molecules in orange and yellow, respectively (2ZVO). (g) Structure of the NZF domain of HOIL-1L (blue) in complex with diubiquitin, showing the additional contacts made by the C-terminal helical extension (3B08).
The donor ubiquitin is held in place by RING2 through contacts that primarily involve hydrophobic interactions with the extended C-terminal tail of ubiquitin. Further stabilization of this arrangement is provided by a β-hairpin located within the helical base of the LDD that is specific to HOIP and together with RING2 guides the C-terminal carboxylate of Gly 76 towards the active-site cysteine of HOIP. The hydrophobic residues of RING2 contacting the donor ubiquitin are conserved in RBRs and may be a general feature of donor ubiquitin recognition by this E3 family. Remarkably, engagement of the C-terminal tail of the thioester-forming ubiquitin in an elongated conformation is also seen in HECT E3 ligases [41,42] and in E2 ∼ Ub conjugates bound to RING ligases [43–45], and may be a general mechanism to prime ubiquitin thioester intermediates for their transfer onto an amino group.
RING1 domains of RBR ligases are structurally similar to canonical RING domains and act to recognize the E2 ∼ Ub conjugate. However, recent work by Klevit and co-workers has uncovered that unlike canonical RING domains, which promote a closed conformation of the E2∼Ub conjugate, RING1 domains actively induce an open conformation of the conjugate instead [46]. Stabilization of a closed E2∼Ub conformation is an important mechanism of RING ligases to activate the conjugate for transfer of ubiquitin onto a lysine side chain [43–45]. By contrast, RBR ligases must prevent the conjugate from unproductive ubiquitin discharge onto a lysine residue to allow formation of the E3∼Ub thioester intermediate. This is achieved by inducing an open E2∼Ub conformation which allows transthiolation, an equilibrium reaction, to proceed while simultaneously suppressing reaction with an amino group [46].
Until recently, structural information on entire RBR domains was restricted to their auto-inhibited states, in which access to the E2-binding site on RING1 and/or access to the catalytic cysteine in RING2 is occluded by intramolecular interactions as manifested in HHARI and Parkin [47–50]. First insight into the architecture of an RBR domain in its active, E2∼Ub bound state was provided by the crystal structure of the RBR-LDD fragment of HOIP in complex with UBE2D∼Ub (figure 3a) [37]. This structure unveiled how the E2∼Ub conjugate is held in an open conformation, tightly embraced by the RBR domain, which uses multiple structural elements to contact the ubiquitin molecule and induce a conformation of the RBR in which the E2∼Ub thioester bond is brought into close proximity of the active-site cysteine in RING2. Intriguingly, the active E2∼Ub/HOIP complex in this arrangement is formed by two separate, elongated RBR molecules that contribute one RING1-IBR and RING2-LDD element each. However, this architecture was suggested to be a crystallization artefact that mimics the active state, normally formed by a monomeric RBR domain. The structure revealed some unexpected features that include the presence of an allosteric ubiquitin-binding site and extensive contacts between the E2-conjugating enzyme and RING2 that overlap with the acceptor ubiquitin-binding site. The allosteric ubiquitin is accommodated by the RING1-IBR linker and IBR domain opposite the E2-conjugated ubiquitin. Biochemical experiments indicate that binding of diubiquitin to this site enhances interaction with E2∼Ub and polyubiquitin chain synthesis, suggesting a feed-forward mechanism of linear chain formation, possibly similar to the effect of phosphorylation of ubiquitin and the Parkin UBL domain on Parkin activity [51]. On the other hand, overlapping contact sides of E2 and acceptor ubiquitin on RING2 imply that the growing polyubiquitin chain has to dissociate before another E2 ∼ Ub conjugate can be bound, thereby suppressing processive chain formation (figure 3c). These at-first-glance contradicting features suggest an intricate mechanism for the regulation of linear chain formation that will require further studies to be fully understood.
4. Disassembly of linear ubiquitin chains
All RBR ligases studied so far exist in an auto-inhibited conformation, in which catalytic activity is suppressed [34,35,47–50]. In the case of HHARI and Parkin, upstream signals are required to relieve auto-inhibition. By contrast, HOIP auto-inhibition is released upon complex formation with HOIL-1L and/or SHARPIN, and work by many groups has shown that complex formation is constitutive and that absence of either destabilizes HOIP [21–23]. This is exemplified by mutations in HOIL-1L and SHARPIN that result in loss of protein and cause disease in humans and mice, respectively (see §§6, 8 and 9 for details) [52,53]. These observations imply that LUBAC is constitutively active and that a mechanism must exist to counterbalance continuous linear chain formation, which would otherwise result in excessive signalling. Such a mechanism has been uncovered with the identification of OTULIN as a DUB specific for the hydrolysis of linear polyubiquitin chains [24,25]. OTULIN was identified independently by two different groups, in one case during the study of mutations in mice that cause embryonic angiogenic deficits [24] and in the other during a bioinformatics screen for unannotated OTU domains (figure 3d) [25]. OTULIN contains the catalytic triad consisting of Cys, His and Asn present in OTU members but unlike other family members is unable to hydrolyse isopeptide bonds and is instead highly specific for cleaving the peptide bond of linear polyubiquitin chains. Linkage specificity is achieved by two key features: the highly selective binding of linear diubiquitin, which is 100-fold tighter than for structurally similar Lys 63-linked diubiquitin, and from substrate-assisted catalysis, in which a residue from the proximal ubiquitin, Glu 16, contributes to the formation of the active site [25]. In the apo state of OTULIN, His 339 of the catalytic triad is contacted by a non-catalytic Asn and prevented from adopting a conformation in which it could deprotonate the catalytic cysteine. Recognition of linear diubiquitin occurs with high affinity (a KD of 150 nM) and involves extensive contacts of OTULIN with conserved surface patches on the distal (Ile 36 and Ile 44 patches) and proximal (Phe 4 patch) ubiquitin molecules. Importantly, the proximal ubiquitin is bound in a conformation that allows its Glu 16 to be inserted into the active site to correctly position His 339 and allow linear ubiquitin chain hydrolysis to occur.
Unlike OTULIN, which is highly specific for linear ubiquitin chains, CYLD is a dual-specificity DUB that cleaves both linear and the structurally similar Lys 63-linked chains [54]. It belongs to the ubiquitin-specific protease (USP) family of DUBs but has a number of insertions and deletions, making it unique within this family. The structures of the USP of zebrafish CYLD in complex with linear (figure 3e) and Lys 63-linked diubiquitin show that the distal ubiquitin is bound in a similar manner in both structures that includes contacts of Leu 8 of the distal ubiquitin with a hydrophobic pocket of CYLD and extensive hydrogen bonds with the extended C-terminal tail of ubiquitin [55]. By contrast, the proximal ubiquitin moieties of linear and Lys 63 diubiquitin are recognized differently by CYLD with an approximately 13° rotation between the two forms. The β9–β10 sheet of CYLD, which is unique to CYLD, interacts with the Phe 4 centred hydrophobic patch of the proximal ubiquitin in both linkage types, while additional contacts specific to either chain type further stabilize the complex. The interaction with both the distal and proximal ubiquitin distinguishes CYLD from other, linkage-independent USP family members, and together with the flexible accommodation of the proximal ubiquitin explains how it is capable of simultaneously recognizing structurally similar linear and Lys 63 chains.
5. Linear ubiquitin chain recognition
Different types of polyubiquitin chains are recognized by chain linkage-specific UBDs to read the signal and translate recognition into the appropriate physiological outcome [56]. UBDs are structurally diverse and no consensus ubiquitin-binding motif exists. Similarly, UBDs contact different surfaces within ubiquitin chains to provide linkage-specific recognition. This may include recognition of surfaces on adjacent ubiquitin molecules in the chain that are only accessible in a specific linkage type, or direct recognition of the bond connecting two ubiquitin molecules. The first UBD with linear chain specificity identified was NEMO/I-κB kinase subunit gamma (IKKγ), the regulatory subunit of the IKK complex [57]. The region recognizing linear ubiquitin chains was termed the ubiquitin binding in ABIN and NEMO, also called the CoZi domain (UBAN). Structural studies of the NEMO UBAN domain bound to linear diubiquitin showed that linear chain specificity relies on specific contacts between NEMO and the distal and proximal ubiquitin, yet that no direct contacts with the peptide bond connecting the two ubiquitin molecules are made (figure 3f) [57]. Interestingly, the specificity of the UBAN domain for linear chains (1.4 µM versus 131 µM for Lys 63 chains) [58,59] is not maintained within the full-length protein where an additional ubiquitin-binding ZF at the C-terminus of NEMO increases the affinity for Lys 63-linked chains considerably [60]. Optineurin/FIP2/NRP, another protein with a UBAN domain, is a suppressor of NF-κB activity due to its ability to compete with NEMO function [61]. It shows high sequence homology with NEMO and its UBAN domain recognizes linear ubiquitin chains in a manner that is highly similar to the interaction between NEMO and linear diubiquitin [62]. Recently, the crystal structure of another UBAN/linear ubiquitin complex has been reported, that between A20-binding inhibitor of NF-κB activation 2 (ABIN2) and linear triubiquitin [63]. While there are similarities in the mechanism of linear diubiquitin recognition to other UBAN domains, there are distinct differences. Most interestingly, the ABIN2–linear triubiquitin interaction occurs with a 2 : 1 stoichiometry in an arrangement that two ABIN2 molecules are bridged by one triubiquitin chain with the middle ubiquitin molecule in the trimer simultaneously contacting two UBAN domains. This unusual geometry suggests a model for the assembly of higher-order signalling complexes by longer polyubiquitin chains that may not only apply to ABIN2 but might be a more general model for the recognition of longer polyubiquitin chains.
Another UBD with specificity for linear ubiquitin chains is found in the DUB A20, which, in addition to the catalytic OTU domain, contains seven ZF domains. Some of these ZFs bind ubiquitin chains of varying topology, whereas others act as protein–protein interaction modules. ZF7 of A20 shows high specificity for linear chains, which is believed to protect them from removal by DUBs, which may explain its apparently opposing effect to CYLD on NF-κB activation [64,65].
Two of the LUBAC subunits, HOIL-1L and SHARPIN, contain NZF domains that recognize linear ubiquitin chains [21,22]. The interaction of the HOIL-1L NZF with linear diubiquitin differs from other NZF–ubiquitin complexes and involves a conserved sequence C-terminal to the NZF domain that adopts an α-helix and contacts the proximal ubiquitin to increase affinity of the interaction (figure 3g) [31]. This feature is unique to the HOIL-1L interaction. The conserved T-F/ϕ motif of the NZF contacts the hydrophobic Ile 44 patch of the distal ubiquitin, while the proximal ubiquitin uses its Phe 4 surface to contact the NZF but no contacts are made between the NZF and the linear linkage itself. Instead, linear chain specificity is achieved by contacting regions of ubiquitin that are only accessible in the linear diubiquitin conformation. At present it is not clear what the physiological relevance of the interaction of LUBAC components with linear ubiquitin chains is. It is tempting to speculate that they may play a regulatory role during chain synthesis, though no evidence for such a behaviour exists at present. Alternatively, they may help to anchor LUBAC to sites where linear chain synthesis is required and thereby help to stabilize macromolecular signalling assemblies.
6. Linear ubiquitin chains in the regulation of immune and cell death signalling pathways
6.1. Tumour necrosis factor cell signalling
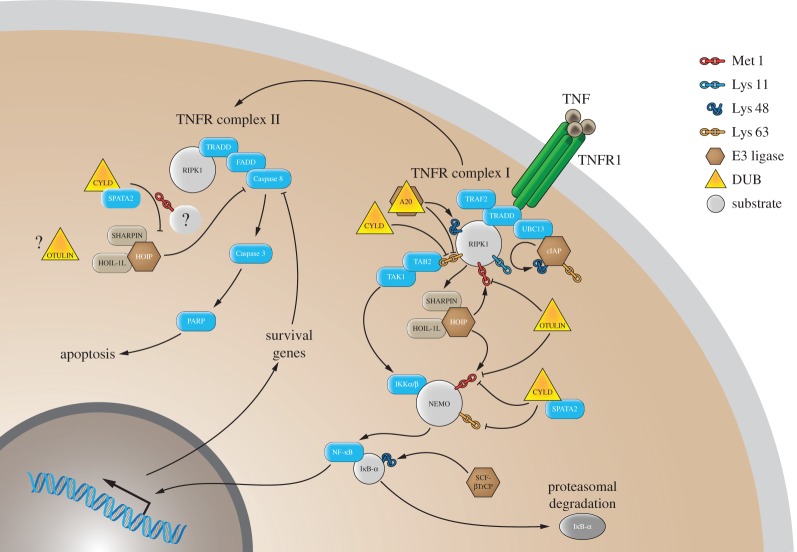
One of the first signalling cascades found to be regulated by LUBAC and linear ubiquitin chains is the tumour necrosis factor (TNF)-signalling pathway (figure 4) [3,4,57]. Upon TNF stimulation, the trimeric TNF receptor (TNFR) recruits a number of signalling molecules including TNFRSF1A associated via death domain (TRADD), TNF receptor associated factor 2 (TRAF2), receptor-interacting protein kinase 1 (RIPK1) and cellular inhibitor of apoptosis protein 1/2 (cIAP1/2). The recruitment of LUBAC components, HOIP, HOIL-1L and SHARPIN to the TNFR complex I has been shown by Walczak and co-workers using moTAP-TNF pull-down assays followed by mass spectrometry analysis [4,22]. In this signalling pathway, LUBAC linearly ubiquitinates its substrates, NEMO and RIPK1 [3,22]. Once linear ubiquitin chains are formed by LUBAC, the NEMO-UBAN domain (see §5) interacts with the chains and subsequently activates NF-κB [57]. In addition to NF-κB activation, LUBAC components HOIL-1L and SHARPIN regulate the TNF-induced JUN N-terminal kinase (JNK) pathway. It was shown that TNF-induced JNK activation is suppressed in HOIL-1L knockdown MCF-7 cells and SHARPIN-deficient chronic proliferative dermatitis mutation (Cpdm) mouse embryonic fibroblasts (MEFs) [4,21,22]. Thus far, substrates of LUBAC regulating JNK are unknown and the detailed mechanisms by which JNK activation is mediated by linear ubiquitination require further studies.
Figure 4.
TNF-induced canonical NF-κB and apoptosis pathways. Different linkage types of ubiquitin chains, Met 1-, Lys 11-, Lys 48- and Lys 63-linked ubiquitin chains, are involved in the TNF-induced canonical NF-κB signalling pathway and the TNFR complex II-dependent apoptosis pathway. Ubiquitin chains of different linkage types are generated by E3 ligases (shown in brown), such as cIAP, HOIP-containing LUBAC complex and SCF-βTrCP. These ubiquitin chains are hydrolysed by DUBs (in yellow) such as OTULIN and CYLD. A20 has a dual role as an E3 ligase and a DUB. Ubiquitination of the substrates including cIAPs, RIPK1, NEMO and IκB-α are critical for the signalling pathway. The TNFR complex II-mediated apoptosis pathway includes RIPK1, TRADD, FADD and Caspase 8. Activation of Caspase 8 leads to the Caspase 3-dependent cleavage of PARP and apoptosis. The LUBAC complex (HOIP, SHARPIN and HOIL-1L) and the CYLD-SPATA2 complex regulates the TNFR complex II-induced apoptosis pathway. Involvement of OTULIN in this apoptosis signalling pathway is not yet determined.
In the TNF signalling cascade, deubiquitinases such as CYLD, OTULIN and A20 [25,29,30,32,33,64,65] negatively regulate NF-κB activation. As described in §4, both OTULIN and CYLD form a complex with the HOIP-PUB domain and this interaction is important for the negative regulation of the NF-κB signalling pathway [26]. When a HOIP-PUB mutant, which is not able to interact with these DUBs, is reconstituted in HOIP-null cells, TNF-induced NF-κB activation is up-regulated [32], suggesting that both OTULIN and CYLD are counterbalancing LUBAC-dependent activation of downstream signalling by forming a complex with HOIP. Importantly, the HOIP-SPATA2-CYLD complex is formed at the TNFR complex, whereas OTULIN does not translocate with LUBAC to the TNFR complex [26,29,33]. As both CYLD and OTULIN negatively regulate the NF-κB signalling cascade, the impact of these deubiquitinases in different LUBAC-dependent signalling cascades may be context-dependent. More recent studies have shown the importance of OTULIN in mice. Immune cell-specific depletion of OTULIN leads to cell-type specific effects: constitutive active NF-κB signalling and overproduction of cytokines in myeloid cells, and down-regulation of linear ubiquitination signal in the B and T cells based on LUBAC degradation [66]. OTULIN and CYLD were also shown to be important regulators of the muramyl dipeptide (MDP)-induced innate immune signalling cascade mediated through RIPK2-ubiquitination [27]. Among the DUBs regulating the NF-κB signalling pathway, A20, which is also known to have E3 ligase activity, has a linear ubiquitin chain interaction domain outside of its catalytic region (as described in §5). The A20-ZF7 interaction with linear Ub chains is crucial for the recruitment of A20 in the TNFR complex, and regulation of TNF-dependent NF-κB activation [64,65]. However, A20 regulates this signalling pathway through its interaction with linear ubiquitin chains and not via its catalytic activity, which distinguishes it from CYLD and OTULIN and implies that its effect is dependent on LUBAC-generated linear ubiquitin chains.
Collectively, LUBAC components, DUBs including OTULIN, CYLD and A20, and linear ubiquitination are critical in many immune response pathways.
6.2. Cell death signalling
Other cell signalling cascades that are regulated by LUBAC are cell death pathways [7,10,67,68]. SHARPIN-deficient Cpdm MEFs are significantly susceptible to TNF-dependent induction of apoptosis (figure 4) [21,22], and TNF-treated HOIL-1L-deficient mice revealed that apoptosis in liver is up-regulated [3]. Furthermore, recent studies both in vivo and in vitro showed that HOIP also plays a critical role in the regulation of apoptosis [69–74]. While substrates for linear ubiquitination during apoptosis signalling remain unclear, it has been established that each of the LUBAC components and LUBAC catalytic activity is required for its anti-apoptosis function [69]. More recent work revealed that HOIP is cleaved by effector Caspases (Caspase 3 and Caspase 6) upon TNF stimulation [75]. Cleavage sites predicted are Asp 348, Asp 387 and Asp 390, resulting in an N-terminal fragment containing PUB-ZF-NZF1 and a C-terminal fragment containing NZF2-UBA-RBR-LDD. The authors demonstrated that cells expressing cleavage-resistant HOIP mutant are protected from TNF combined with cycloheximide (CHX)-induced apoptosis. The remaining question is whether TNF-induced apoptosis by HOIP cleavage involves a linear ubiquitination event. The SPATA2-CYLD complex also plays a role in the TNF-dependent apoptosis pathway [76]. Different studies highlighted that TNF-induced apoptosis due to LUBAC deficiency is dependent on the TNFR complex II pathway (figure 4) [10,67,68]. For example, it was shown that targeting TNFR complex II components by the expression of dominant negative FADD or Caspase 8 inhibitor, CrmA in Cpdm MEFs inhibits apoptosis, suggesting that apoptosis is mediated through the TNFR complex II [21].
Cpdm mice, which have a nonsense mutation in the SHARPIN gene, suffer from systemic inflammation [53]. In these mice, secondary lymphoid organ development and differentiation of T cells and B cells are defective, leading to deficiency of IgG production [53]. Especially the skin inflammation phenotype accompanied with apoptosis observed in Cpdm mice is largely rescued by epithelial-specific TRADD deficiency, and epithelial-specific Fas-associated protein with death domain (FADD) and RIPK3 deficiency [70,71]. These observations suggest that apoptosis plays a major role in the regulation of skin inflammation caused by SHARPIN deficiency. On the other hand, because deficiency of the necroptosis regulators RIPK3 and mixed lineage kinase domain-like protein (MLKL) had only mild effects on skin inflammation, necroptosis has a minor role in this model. The skin inflammatory phenotype and induction of cell death in skin tissue observed in Cpdm mice was largely rescued by double genetic ablation of Caspase 1 and Caspase 11, whereas inflammation in other organs remained [77]. An independent study revealed functional differences between Caspase 1 and Caspase 11 by using Caspase 1-deficient (Caspase 1−/−; Caspase 11 transgenic) mice, showing that Caspase 1 has a major role in the regulation of inflammatory phenotype in Cpdm mice [78]. Mechanistically, SHARPIN directly interacts with Caspase 1 and negatively regulates the activity of Caspase 1, leading to the suppression of mature IL-1β and IL-18 production. Whether the role of SHARPIN in Caspase 1/11-dependent skin inflammation depends on the catalytic function of LUBAC remains to be understood. The cross of HOIL-1-L-deficient mice with Cpdm mice leads to embryonic lethality at the embryonic stage E10.5 accompanied with up-regulation of apoptosis, identical to HOIPΔlinear/Δlinear mice in which the catalytic RBR region is deleted [79]. These observations suggest cooperative roles of HOIL-1-L and SHARPIN in mouse embryonic development and apoptosis. Similar to HOIPΔlinear/Δlinear mice, HOIP-deficient mice and HOIP catalytic inactive mutant knockin mice are embryonic lethal at E10.5 [40,69], indicating a critical role of HOIP catalytic activity in regulation of mouse embryonic development and apoptosis.
The role of LUBAC in the cell death signalling cascade is not limited to the TNF-induced pathway; Walczak and co-workers have shown both in vitro and in vivo that polyI:C-induced Toll-like receptor 3 (TLR3)-dependent apoptosis also involves LUBAC [80]. It was also demonstrated that HOIP plays an anti-apoptosis role in the cisplatin-induced genotoxicity pathway via the protein kinase ataxia–telangiectasia mutated (ATM) [72].
These observations collectively indicate a critical role of LUBAC and linear ubiquitination in the regulation of various apoptosis signalling pathways.
6.3. Other immune signalling pathways
It is well established by now that LUBAC plays an important role in multiple types of immune responses, including activation of NF-κB induced by various ligands, such as peptidoglycan (PGN), CD40 ligand (CD40-L), interleukin 1beta (IL-1β) and lipopolysaccharide (LPS) in B cells or in macrophages [21,22,81,82]. For example, PGN-dependent induction of the nucleotide-binding oligomerization domain-containing protein 1/2 (NOD1/2) pattern recognition receptor signalling cascade leads to linear ubiquitination of RIPK2 by recruiting LUBAC into the receptor complex [81,82]. In this signalling cascade, LUBAC ligase activity is required for efficient NF-κB activation and secretion of pro-inflammatory cytokines. As previously described (in §3), Lys 63–Met 1 hybrid ubiquitin polymers are formed on the substrates, such as NEMO in the IL-1β immune signalling cascades, RIPK2 via the NOD2 pathway and RIPK1 via TLR3 or TNFR [40,83]. These are very interesting observations suggesting that LUBAC-induced linear ubiquitination may depend on the initial Lys 63 ubiquitin chain formation on the substrates.
LUBAC is also involved in the regulation of TLR signalling cascades. One of the first studies that linked TLR signalling and SHARPIN was based on a system-level analysis of TLR-stimulated macrophages [84]. The authors demonstrated that macrophages derived from Cpdm mice were defective in IL-12 production induced by TLR activation, and that the effects of SHARPIN deficiency on the TLR2-induced transcriptome were highly correlated with the effects of the hypomorphic Leu 153 Pro/panr2 point mutation in the gene encoding NEMO. Furthermore, HOIP was demonstrated to be involved in the TLR3-induced signalling cascade examined in HOIP-deficient HaCaT or MEFs [80,83], as well as HOIL-1L in the TLR4 signalling pathway using an LPS-induced systemic inflammation model in HOIL-1L-deficient mice [85] and in a mouse B-cell line [86].
Additional roles for LUBAC during innate immune signalling have been described including NEMO ubiquitination in the MAVS-TRAF3 complex [87], IRF3 ubiquitination in the RLR-induced IRF-3-mediated pathway of apoptosis (RIPA) [88] and ASC ubiquitination in the NLRP3/ASC inflammasome signalling pathway [85]. Furthermore, it has been suggested that LUBAC plays a role in LMP1 signalling [89], and in the RIG-I TRIM25-mediated IFN pathway [90]; however, it is not yet clear at present if this requires LUBAC catalytic activity, and the mechanistic understanding of LUBAC function in these pathways awaits further studies. Recent work from multiple groups showed an important function of mucosa associated lymphoid tissue lymphoma translocation gene 1 (MALT1)-dependent HOIL-1L cleavage in the negative regulation of the T-cell receptor (TCR)-induced NF-κB signalling cascade [91–94]. The MALT-1-cleaved products of HOIL-1L are an N-terminal fragment containing the UBL, and a C-terminal fragment containing the NZF and RBR domains. Although both of these HOIL-1L protein fragments are stable in cells, cleavage leads to destabilization of HOIP [91]. This is rather surprising because the HOIL-1L UBL domain is sufficient to interact with HOIP. Based on these studies, we speculate that there might be an additional interplay between HOIP and HOIL-1L that is not understood yet. In the TCR signalling pathway, it was also shown that the B-cell lymphoma/leukaemia 10 (Bcl10) is linearly ubiquitinated and recognized by NEMO. Caspase recruitment domain family member 11 (CARD11) and MALT1 are both required for the TCR-induced linear ubiquitination of Bcl10 as demonstrated in CARD11- and MALT1-deficient Jurkat T cells [95]. To fully understand the functional relationship between MALT1-dependent HOIL-1-L cleavage and subsequent inactivation of NF-κB signalling and Bcl10 ubiquitination, further studies are required.
LUBAC plays a critical role in late thymocyte differentiation, FOXP3+ regulatory T (Treg)-cell development and Treg cell homeostasis [96]. Treg-specific HOIP ablation leads to lethal immune pathology in mice [96]. Furthermore, the HOIP-RBR region regulates T-cell and B-cell differentiation. In T-cell-specific HOIPΔlinear mice, in which HOIP-RBR is deleted, thymic CD4+ or CD8+ T-cell numbers were markedly reduced with severe defects in NKT cell development [73]. In B cells derived from B-cell-specific HOIPΔlinear mice, the development of the B1 cells is defective and CD40- or TACI-induced canonical NF-κB and ERK signalling pathways are impaired [97]. Collectively, these studies show that LUBAC plays an important role in adaptive immunity.
7. Linear ubiquitination in the non-immune signalling cascades
In addition to its well-established role in the regulation of immune signalling and apoptosis (as described in §6), it is becoming apparent that linear ubiquitin chains also have other cellular roles.
7.1. Wnt signalling pathway regulated by OTULIN
Linear ubiquitination regulators are involved in non-immune signalling cascades as illustrated in Gumby mice, in which the OTULIN gene has missense mutations that are embryonic lethal due to vascular formation defects [24]. The interaction of OTULIN with a key component Dishevelled (Dvl/Dsh) mediates this regulation through the Wnt signalling cascade.
7.2. Genotoxic stress-induced NF-κB activation by LUBAC
The LUBAC components were shown to be important for NF-κB activation induced by genotoxic stress [98]. Genotoxic stress in cells leads to linear ubiquitination of NEMO in the cytosolic fraction, which requires LUBAC, and subsequent activation of NF-κB. The molecular mechanism that mediates LUBAC activation upon genotoxic stress remains to be understood.
7.3. Linear ubiquitination by Drosophila linear ubiquitin E3 ligase in heat tolerance
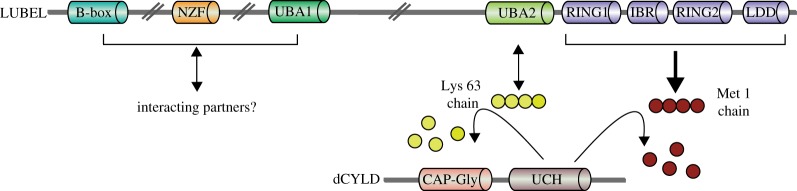
More recently, a ubiquitin E3 ligase in Drosophila, which specifically generates linear ubiquitin chains, was identified [99]. Drosophila linear ubiquitin E3 ligase (LUBEL)/CG11321 consists of a highly conserved RING1-RBR-RING2-LDD catalytic domain, and protein–protein interaction domains including a B-box, NZF and two UBAs (figure 5). In comparison with human HOIP, the N-terminal region of LUBEL is stretched out with large insertions between recognizable domains, and LUBEL is approximately three times larger. Furthermore, some of the protein–protein interaction domains present in human HOIP (PUB, ZF and one of the NZF domains) are not present in LUBEL. Based on bioinformatic analysis, genes for the LUBAC subunits, namely SHARPIN or HOIL-1L, and the linear ubiquitin-specific DUB, OTULIN, are not predicted in the Drosophila genome. Even without these subunits, LUBEL alone generates linear ubiquitin chains in a RING/HECT hybrid manner. The DUB for linear chains in Drosophila is dCYLD, which was confirmed to cleave linear and Lys 63-linked ubiquitin chains, similar to human CYLD. In LUBEL, the PUB domain, which is responsible for recruiting CYLD and SPATA2 in human HOIP, is lacking. Instead, the LUBEL C-terminal region (the RBR-LDD domain) interacts with recombinant dCYLD in vitro. An orthologue of SPATA2 in Drosophila called Tamo was shown to have a conserved PUB domain [100] and it is of high interest to understand if Tamo is involved in the regulation of linear ubiquitination in Drosophila. These observations suggest that, in Drosophila, LUBEL and dCYLD play an important role in the regulation of linear ubiquitin chain formation and destruction. In contrast with HOIP knockout or catalytic inactive knockin mice that are embryonic lethal, catalytic inactive knockin LUBEL flies are viable and showed no obvious developmental defect. LUBEL knockdown flies and LUBEL catalytic inactive mutant flies, in which linear ubiquitination is below the detection limit, are defective in heat tolerance, which may be relevant also in the mammalian system.
Figure 5.
Domain structures of Drosophila LUBEL and dCYLD. Drosophila LUBEL-RBR-LDD is the catalytic region for linear ubiquitination. The UBA2 domain interacts with Lys 63-linked ubiquitin chains. An N-terminal region of LUBEL may interact with unknown interacting partners. UBA2 interacts with Lys 63-linked ubiquitin chains and dCYLD cleaves linear and Lys 63-linked ubiquitin chains. Npl4 zinc finger (NZF), ubiquitin (Ub)-associated (UBA1 and UBA2), RING between RING (RBR)-C. The RBR-C region consists of RING1, in-between RING (IBR), RING2 and linear Ub chain-determining domain (LDD), cytoskeletal-associated protein-glycine-conserved (CAP-Gly) domain and ubiquitin carboxyl-terminal hydrolase (UCH). The double bars between domains indicate that they are separated by large stretches of sequence, resulting in LUBEL being almost three times larger (2892 amino acids) than human HOIP (1072 amino acids).
These observations suggest that there is more to learn about the roles of LUBAC and linear ubiquitination in the mammalian system in addition to their roles in immune and apoptotic signalling.
8. The role of the LUBAC component SHARPIN independent of catalytic activity
The LUBAC subunit, SHARPIN was shown to interact with integrin and inhibit β-integrin activity [101]. This interaction is mediated by the SHARPIN UBL domain and important residues for the interaction with integrin are shared with the HOIP interaction [102]. Because LUBAC forms a relatively stable complex and depletion of a LUBAC component in cells often leads to destabilization of the other complex components, it will be interesting to understand how the integrin–SHARPIN complex is formed, and how SHARPIN may determine its interacting partner in cells, namely HOIP or integrin. Recently, it was demonstrated that SHARPIN controls mouse mammary gland development [103]. Whether these events depend on the ubiquitination function of the LUBAC complex or alternatively depend on the role of SHARPIN alone remains to be understood.
9. Linear ubiquitin chains and disease
9.1. Human diseases associated with mutations in HOIP and HOIL-1L
In a patient with multi-organ auto-inflammation, combined immunodeficiency, subclinical amylopectinosis and systemic lymphangiectasia, the HOIP gene is homozygous for a mutation at Leu 72 Pro (L72P) in the PUB domain (described in §2) [104] (table 1). The HOIP L72P mutation leads to destabilization of HOIP itself and LUBAC. Both IL-1β and TNF-induced NF-κB activation and linear ubiquitination are impaired in fibroblasts derived from the patient. However, the patient's monocytes respond to IL-1β more vigorously than control monocytes, whereas the activation and differentiation of the patient's B cells are impaired in response to CD40 engagement. These cellular and clinical phenotypes largely overlap those of HOIL-1L-deficient patients. For example, it was reported that a new fatal human inherited disorder characterized by chronic auto-inflammation, invasive bacterial infections and muscular amylopectinosis carried biallelic loss-of-expression and loss-of-function mutations in HOIL-1L (RBCK1) [52]. Moreover, RBCK1 gene mutations were identified in non-immune disease patients, namely in glycogen storage disease patients, who suffer from myopathy and cardiomyopathy. It was found that 10 patients from eight different families had heterozygous missense or truncating mutations in the RBCK1 gene [105,106]. These observations suggest a role of HOIL-1L in the regulation of non-immune pathology and cellular functions, which are independent of immune system-dependent NF-κB activation.
HOIP has also been shown to be involved in activated B-cell-diffuse large B-cell lymphoma (ABC-DLBCL). Two germ-line polymorphisms affecting HOIP are rare among healthy individuals but enriched in ABC-DLBCL. These are located in the UBA of HOIP that mediates the interaction with HOIL-1-L (see §2), and have been suggested to increase their association, thereby leading to up-regulation of NF-κB [109]. Activation of IKK-dependent NF-κB through the CARD11-MALT1-BCL10 (CBM) complex in the chronic active B-cell receptor (BCR) signalling cascade, which is a hallmark of ABC-DLBCL, includes the ubiquitin E3 ligases cIAP1, cIAP2 as well as LUBAC. cIAP1/2-induced Lys 63 ubiquitin chains on cIAPs and Bcl10 recruits LUBAC to the signalling complex, which is crucial for subsequent IKK activation [108]. This aligns with the previous finding in the TNFR signalling cascade that the E3 ligase activity of cIAPs is required for LUBAC recruitment into the receptor complex.
9.2. OTULIN-related auto-inflammatory syndrome
Two independent groups reported that homozygous gene mutations of OTULIN/FAM105B were found in autoimmune disease patients [66,107] (table 1). Loss of function mutations or a missense mutation in human OTULIN causes a potentially fatal auto-inflammatory condition, which was termed OTULIN-related auto-inflammatory syndrome (ORAS). Expression of immunologically related genes in whole blood and fibroblasts derived from patients was high in comparison to the cells from a healthy patient. The mutations found in these patients, Tyr 244 Cys and Leu 272 Pro, are located near or on the S1 distal ubiquitin binding site [107]. It was confirmed that the Leu 272 Pro mutation abolishes the catalytic activity of OTULIN [66].
Table 1.
Reported mutations in RNF31, RBCK1 and FAM105B genes in human patients.
| gene name | protein name | gene mutations (amino acid alterations) | protein domains affected | symptoms | reference |
|---|---|---|---|---|---|
| RNF31 | HOIP | L72P (homozygous) | PUB | multi-organ auto-inflammation, combined immunodeficiency, subclinical amylopectinosis, and systemic lymphangiectasia | [104] |
| RBCK1 | HOIL-1L | L41fsX7 (homozygous) | all domains (a mutation at the N-terminus to UBL) | chronic auto-inflammation, invasive bacterial infections and muscular amylopectinosis | [52] |
| Q185X;c.ex1_ex4del (compound heterozygous) | NZF-RING1-IBR-RING2 (a mutation at the N-terminus to NZF); UBL (deletion of 1-154) | chronic auto-inflammation, invasive bacterial infections and muscular amylopectinosis | [52] | ||
| E243X; N387S (compound heterozygous) | RING1-IBR-RING2 (a mutation between NZF and RING1); IBR | myopathy and cardiomyopathy | [105] | ||
| E299VfsX18 (homozygous) | RING1-IBR-RING2 (a mutation within RING1) | myopathy and cardiomyopathy | [105] | ||
| A241GfsX34 (homozygous) | RING1-IBR-RING2 (a mutation between NZF and RING1) | myopathy and cardiomyopathy | [105] | ||
| A18P (homozygous) | N-terminus | myopathy and cardiomyopathy | [105] | ||
| E243GfsX114; c.ex1_ex4del (compound heterozygous) | RING1-IBR-RING2(a mutation between NZF and RING1); UBL (deletion of 1-154) | myopathy and cardiomyopathy | [105] | ||
| R352X (homozygous) | IBR | myopathy and cardiomyopathy | [105] | ||
| R298RfsX40 (homozygous) | RING1 | myopathy and cardiomyopathy | [105] | ||
| R165RfsX111 (homozygous) | NZF-RING1-IBR-RING2 (a mutation between UBL and NZF) | myopathy and cardiomyopathy | [105] | ||
| Q222X;E190fs (compound heterozygous) | NZF-tail-RING1-IBR-RING2 (a mutation between NZF and RING1); NZF-RING1-IBR-RING2 | progressive muscular weakness and cardiomyopathy | [106] | ||
| FAM105B | OTULIN | L272P (homozygous) | OTU | auto-inflammatory syndrome | [66,107] |
| Y244C (homozygous) | OTU | auto-inflammation, paniculitis and dermatosis | [107] | ||
| G174DfsX2 (homozygous) | OTU | auto-inflammation, paniculitis and dermatosis | [107] |
9.3. Lupus/autoimmune diseases, the E2 enzyme UBE2L3 and linear ubiquitin chains
There are several reports linking a single risk haplotype across the UBE2L3/UBCH7 gene with systemic lupus erythematosus and autoimmune diseases [110,111]. UBE2L3/UBCH7 specifically acts with HECT-type and RBR family E3 ubiquitin ligases including HOIP and HOIL-1L. Based on a single risk haplotype across UBE2L3, it was hypothesized that UBE2L3 together with LUBAC regulates NF-κB. Indeed, it was shown that UBE2L3 is the preferred E2-conjugating enzyme for LUBAC in vivo, and that UBE2L3 is essential for LUBAC-mediated activation of NF-κB. How LUBAC recognizes UBE2L3 over other E2s such as UBE2D is yet to be clarified.
10. Conclusion
Since the first biochemical study of LUBAC-induced linear ubiquitination was reported by Iwai and co-workers in 2006 [2], our understanding of LUBAC and linear ubiquitination has expanded significantly from molecular and cellular functions to its impact on in vivo roles and human diseases. Linear ubiquitin chains belong to those polyubiquitin chain types that are not primarily used for proteasomal degradation of proteins but rather used as signalling tags to increase the functional variety in regulatory mechanisms of cellular responses. Understanding the molecular specificity of linear ubiquitination, namely (i) how and which enzymes generate or disassemble the chains, (ii) how substrates are chosen for linear ubiquitination, and (iii) how linear ubiquitin chains are recognized, will allow a better dissection of the biology regulated by linear ubiquitination. Great progress has been made in recent years with the discovery of novel regulators of LUBAC, the finding that ubiquitin chains can be formed as hybrids of linear and Lys 63 chains and the association of mutations in LUBAC with human disease, and it will be exciting to see where the linear ubiquitin chain will take us next.
Acknowledgements
We thank Jorge Almagro for discussions and critical reading of the manuscript, and the graphic departments at the Crick Institute and the IMP/IMBA for their support in generating figures. We apologize to the authors whose papers we could not cite due to the space limitations.
Competing interests
We declare we have no competing interests.
Funding
F.I. is supported by the ERC consolidator grant (LUbi, 614711), the FWF standalone grant (P 25508), COST (European Cooperation in Science and Technology, PROTEOSTASIS BM1307) and Austrian Academy of Sciences, and K.R. by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001142), the UK Medical Research Council (FC001142) and the Wellcome Trust (FC001142).
References
- 1.Komander D, Rape M. 2012. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229. (doi:10.1146/annurev-biochem-060310-170328) [DOI] [PubMed] [Google Scholar]
- 2.Kirisako T, et al. 2006. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887. (doi:10.1038/sj.emboj.7601360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tokunaga F, et al. 2009. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat. Cell Biol. 11, 123–132. (doi:10.1038/ncb1821) [DOI] [PubMed] [Google Scholar]
- 4.Haas TL, et al. 2009. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell. 36, 831–844. (doi:10.1016/j.molcel.2009.10.013) [DOI] [PubMed] [Google Scholar]
- 5.Fiil BK, Gyrd-Hansen M. 2014. Met1-linked ubiquitination in immune signalling. FEBS J. 281, 4337–4350. (doi:10.1111/febs.12944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikeda F. 2015. Linear ubiquitination signals in adaptive immune responses. Immunol. Rev. 266, 222–236. (doi:10.1111/imr.12300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwai K, Fujita H, Sasaki Y. 2014. Linear ubiquitin chains: NF-κB signalling, cell death and beyond. Nat. Rev. Mol. Cell Biol. 15, 503–508. (doi:10.1038/nrm3836) [DOI] [PubMed] [Google Scholar]
- 8.Shimizu Y, Taraborrelli L, Walczak H. 2015. Linear ubiquitination in immunity. Immunol. Rev. 266, 190–207. (doi:10.1111/imr.12309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elton L, Carpentier I, Verhelst K, Staal J, Beyaert R. 2015. The multifaceted role of the E3 ubiquitin ligase HOIL-1: beyond linear ubiquitination. Immunol. Rev. 266, 208–221. (doi:10.1111/imr.12307) [DOI] [PubMed] [Google Scholar]
- 10.Sasaki K, Iwai K. 2015. Roles of linear ubiquitinylation, a crucial regulator of NF-κB and cell death, in the immune system. Immunol. Rev. 266, 175–189. (doi:10.1111/imr.12308) [DOI] [PubMed] [Google Scholar]
- 11.Hershko A, Ciechanover A. 1998. The ubiquitin system. Ann. Rev. Biochem. 67, 425–479. (doi:10.1146/annurev.biochem.67.1.425) [DOI] [PubMed] [Google Scholar]
- 12.Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434. (doi:10.1146/annurev.biochem.78.101807.093809) [DOI] [PubMed] [Google Scholar]
- 13.Berndsen CE, Wolberger C. 2014. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21, 301–307. (doi:10.1038/nsmb.2780) [DOI] [PubMed] [Google Scholar]
- 14.Buetow L, Huang DT. 2016. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 17, 626–642. (doi:10.1038/nrm.2016.91) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komander D, Clague MJ, Urbe S. 2009. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563. (doi:10.1038/nrm2731) [DOI] [PubMed] [Google Scholar]
- 16.Heideker J, Wertz IE. 2015. DUBs, the regulation of cell identity and disease. Biochem. J. 467, 191 (doi:10.1042/bj4670191) [DOI] [PubMed] [Google Scholar]
- 17.Kim HC, Huibregtse JM. 2009. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29, 3307–3318. (doi:10.1128/MCB.00240-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. 2011. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108. (doi:10.1038/nature09966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smit JJ, Sixma TK. 2014. RBR E3-ligases at work. EMBO Rep. 15, 142–154. (doi:10.1002/embr.201338166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spratt DE, Walden H, Shaw GS. 2014. RBR E3 ubiquitin ligases: new structures, new insights, new questions. Biochem. J. 458, 421–437. (doi:10.1042/BJ20140006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda F, et al. 2011. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471, 637–641. (doi:10.1038/nature09814) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerlach B, et al. 2011. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596. (doi:10.1038/nature09816) [DOI] [PubMed] [Google Scholar]
- 23.Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K. 2011. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471, 633–636. (doi:10.1038/nature09815) [DOI] [PubMed] [Google Scholar]
- 24.Rivkin E, et al. 2013. The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature 498, 318–324. (doi:10.1038/nature12296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keusekotten K, et al. 2013. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell 153, 1312–1326. (doi:10.1016/j.cell.2013.05.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kupka S, De Miguel D, Draber P, Martino L, Surinova S, Rittinger K, Walczak H. 2016. SPATA2-mediated binding of CYLD to HOIP enables CYLD recruitment to signaling complexes. Cell Rep. 16, 2271–2280. (doi:10.1016/j.celrep.2016.07.086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elliott PR, et al. 2016. SPATA2 links CYLD to LUBAC, activates CYLD, and controls LUBAC signaling. Mol. Cell. 63, 990–1005. (doi:10.1016/j.molcel.2016.08.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yagi H, et al. 2012. A non-canonical UBA-UBL interaction forms the linear-ubiquitin-chain assembly complex. EMBO Rep. 13, 462–468. (doi:10.1038/embor.2012.24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elliott PR, Nielsen SV, Marco-Casanova P, Fiil BK, Keusekotten K, Mailand N, Freund SM, Gyrd-Hansen M, Komander D. 2014. Molecular basis and regulation of OTULIN-LUBAC interaction. Mol. Cell. 54, 335–348. (doi:10.1016/j.molcel.2014.03.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaeffer V, Akutsu M, Olma MH, Gomes LC, Kawasaki M, Dikic I. 2014. Binding of OTULIN to the PUB domain of HOIP controls NF-κB signaling. Mol. Cell. 54, 349–361. (doi:10.1016/j.molcel.2014.03.016) [DOI] [PubMed] [Google Scholar]
- 31.Sato Y, Fujita H, Yoshikawa A, Yamashita M, Yamagata A, Kaiser SE, Iwai K, Fukai S. 2011. Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex. Proc. Natl Acad. Sci. USA 108, 20 520–20 525. (doi:10.1073/pnas.1109088108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takiuchi T, et al. 2014. Suppression of LUBAC-mediated linear ubiquitination by a specific interaction between LUBAC and the deubiquitinases CYLD and OTULIN. Genes Cells 19, 254–272. (doi:10.1111/gtc.12128) [DOI] [PubMed] [Google Scholar]
- 33.Draber P, et al. 2015. LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Rep. 13, 2258–2272. (doi:10.1016/j.celrep.2015.11.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stieglitz B, Morris-Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K. 2012. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 13, 840–846. (doi:10.1038/embor.2012.105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smit JJ, Monteferrario D, Noordermeer SM, van Dijk WJ, van der Reijden BA, Sixma TK. 2012. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J. 31, 3833–3844. (doi:10.1038/emboj.2012.217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stieglitz B, et al. 2013. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature 503, 422–426. (doi:10.1038/nature12638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lechtenberg BC, Rajput A, Sanishvili R, Dobaczewska MK, Ware CF, Mace PD, Riedl SJ. 2016. Structure of a HOIP/E2∼ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature 529, 546–550. (doi:10.1038/nature16511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smit JJ, van Dijk WJ, El Atmioui D, Merkx R, Ovaa H, Sixma TK. 2013. Target specificity of the E3 ligase LUBAC for ubiquitin and NEMO relies on different minimal requirements. J. Biol. Chem. 288, 31 728–31 737. (doi:10.1074/jbc.M113.495846) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujita H, Rahighi S, Akita M, Kato R, Sasaki Y, Wakatsuki S, Iwai K. 2014. Mechanism underlying IkB kinase activation mediated by the linear ubiquitin chain assembly complex. Mol. Cell. Biol. 34, 1322–1335. (doi:10.1128/MCB.01538-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emmerich CH, Ordureau A, Strickson S, Arthur JS, Pedrioli PG, Komander D, Cohen P. 2013. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl Acad. Sci. USA 110, 15 247–15 252. (doi:10.1073/pnas.1314715110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamadurai HB, et al. 2013. Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. ELife 2, e00828 (doi:10.7554/eLife.00828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maspero E, Valentini E, Mari S, Cecatiello V, Soffientini P, Pasqualato S, Polo S. 2013. Structure of a ubiquitin-loaded HECT ligase reveals the molecular basis for catalytic priming. Nat. Struct. Mol. Biol. 20, 696–701. (doi:10.1038/nsmb.2566) [DOI] [PubMed] [Google Scholar]
- 43.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. 2012. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120. (doi:10.1038/nature11376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. 2012. Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell. 47, 933–942. (doi:10.1016/j.molcel.2012.07.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. 2012. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 19, 876–883. (doi:10.1038/nsmb.2379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dove KK, Stieglitz B, Duncan ED, Rittinger K, Klevit RE. 2016. Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms. EMBO Rep. 17, 1221–1235. (doi:10.15252/embr.201642641) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riley BE, et al. 2013. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun. 4, 1982 (doi:10.1038/ncomms2982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trempe JF, et al. 2013. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340, 1451–1455. (doi:10.1126/science.1237908) [DOI] [PubMed] [Google Scholar]
- 49.Wauer T, Komander D. 2013. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 32, 2099–2112. (doi:10.1038/emboj.2013.125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duda DM, Olszewski JL, Schuermann JP, Kurinov I, Miller DJ, Nourse A, Alpi AF, Schulman BA. 2013. Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure 21, 1030–1041. (doi:10.1016/j.str.2013.04.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ordureau A, et al. 2014. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell. 56, 360–375. (doi:10.1016/j.molcel.2014.09.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boisson B, et al. 2012. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat. Immunol. 13, 1178–1186. (doi:10.1038/ni.2457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seymour RE, Hasham MG, Cox GA, Shultz LD, Hogenesch H, Roopenian DC, Sundberg JP. 2007. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun. 8, 416–421. (doi:10.1038/sj.gene.6364403) [DOI] [PubMed] [Google Scholar]
- 54.Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, Barford D. 2008. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol. Cell. 29, 451–464. (doi:10.1016/j.molcel.2007.12.018) [DOI] [PubMed] [Google Scholar]
- 55.Sato Y, et al. 2015. Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity. Nat. Struct. Mol. Biol. 22, 222–229. (doi:10.1038/nsmb.2970) [DOI] [PubMed] [Google Scholar]
- 56.Husnjak K, Dikic I. 2012. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322. (doi:10.1146/annurev-biochem-051810-094654) [DOI] [PubMed] [Google Scholar]
- 57.Rahighi S, et al. 2009. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136, 1098–1109. (doi:10.1016/j.cell.2009.03.007) [DOI] [PubMed] [Google Scholar]
- 58.Ivins FJ, Montgomery MG, Smith SJ, Morris-Davies AC, Taylor IA, Rittinger K. 2009. NEMO oligomerization and its ubiquitin-binding properties. Biochem. J. 421, 243–251. (doi:10.1042/BJ20090427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lo YC, Lin SC, Rospigliosi CC, Conze DB, Wu CJ, Ashwell JD, Eliezer D, Wu H. 2009. Structural basis for recognition of diubiquitins by NEMO. Mol. Cell 33, 602–615. (doi:10.1016/j.molcel.2009.01.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laplantine E, Fontan E, Chiaravalli J, Lopez T, Lakisic G, Veron M, Agou F, Israel A. 2009. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 28, 2885–2895. (doi:10.1038/emboj.2009.241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu G, Wu CJ, Zhao Y, Ashwell JD. 2007. Optineurin negatively regulates TNFalpha-induced NF-κB activation by competing with NEMO for ubiquitinated RIP. Curr. Biol. 17, 1438–1443. (doi:10.1016/j.cub.2007.07.041) [DOI] [PubMed] [Google Scholar]
- 62.Nakazawa S, et al. 2016. Linear ubiquitination is involved in the pathogenesis of optineurin-associated amyotrophic lateral sclerosis. Nat. Commun. 7, 12547 (doi:10.1038/ncomms12547) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin SM, Lin SC, Hong JY, Su TW, Kuo BJ, Chang WH, Tu YF, Lo YC. 2017. Structural insights into linear Tri-ubiquitin recognition by A20-binding inhibitor of NF-κB, ABIN-2. Structure 25, 66–78. (doi:10.1016/j.str.2016.11.005) [DOI] [PubMed] [Google Scholar]
- 64.Tokunaga F, et al. 2012. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO J. 31, 3856–3870. (doi:10.1038/emboj.2012.241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Verhelst K, Carpentier I, Kreike M, Meloni L, Verstrepen L, Kensche T, Dikic I, Beyaert R. 2012. A20 inhibits LUBAC-mediated NF-κB activation by binding linear polyubiquitin chains via its zinc finger 7. EMBO J. 31, 3845–3855. (doi:10.1038/emboj.2012.240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Damgaard RB, et al. 2016. The deubiquitinase OTULIN is an essential negative regulator of inflammation and autoimmunity. Cell 166, 1215–1230. (doi:10.1016/j.cell.2016.07.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rieser E, Cordier SM, Walczak H. 2013. Linear ubiquitination: a newly discovered regulator of cell signalling. Trends Biochem. Sci. 38, 94–102. (doi:10.1016/j.tibs.2012.11.007) [DOI] [PubMed] [Google Scholar]
- 68.Asaoka T, Ikeda F. 2015. New insights into the role of ubiquitin networks in the regulation of antiapoptosis pathways. Int. Rev. Cell Mol. Biol. 318, 121–158. (doi:10.1016/bs.ircmb.2015.05.003) [DOI] [PubMed] [Google Scholar]
- 69.Peltzer N, et al. 2014. HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Rep. 9, 153–165. (doi:10.1016/j.celrep.2014.08.066) [DOI] [PubMed] [Google Scholar]
- 70.Kumari S, et al. 2014. Sharpin prevents skin inflammation by inhibiting TNFR1-induced keratinocyte apoptosis. ELife 3, e03422 (doi:10.7554/eLife.03422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rickard JA, et al. 2014. TNFR1-dependent cell death drives inflammation in Sharpin-deficient mice. ELife 3, e03464 (doi:10.7554/eLife.03464) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mackay C, Carroll E, Ibrahim AF, Garg A, Inman GJ, Hay RT, Alpi AF. 2014. E3 ubiquitin ligase HOIP attenuates apoptotic cell death induced by cisplatin. Cancer Res. 74, 2246–2257. (doi:10.1158/0008-5472.CAN-13-2131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okamura K, Kitamura A, Sasaki Y, Chung DH, Kagami S, Iwai K, Yasutomo K. 2016. Survival of mature T cells depends on signaling through HOIP. Sci. Rep. 6, 36135 (doi:10.1038/srep36135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shimizu Y, Peltzer N, Sevko A, Lafont E, Sarr A, Draberova H, Walczak H. In press. The linear ubiquitin chain assembly complex acts as a liver tumor suppressor and inhibits hepatocyte apoptosis and hepatitis. Hepatology (doi:10.1002/hep.29074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Joo D, Tang Y, Blonska M, Jin J, Zhao X, Lin X. 2016. Regulation of linear ubiquitin chain assembly complex by caspase-mediated cleavage of RNF31. Mol. Cell. Biol. 36, 3010–3018. (doi:10.1128/MCB.00474-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schlicher L, Wissler M, Preiss F, Brauns-Schubert P, Jakob C, Dumit V, Borner C, Dengjel J, Maurer U. 2016. SPATA2 promotes CYLD activity and regulates TNF-induced NF-κB signaling and cell death. EMBO Rep. 17, 1485–1497. (doi:10.15252/embr.201642592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Douglas T, Champagne C, Morizot A, Lapointe JM, Saleh M. 2015. The inflammatory caspases-1 and -11 mediate the pathogenesis of dermatitis in Sharpin-deficient mice. J. Immunol. 195, 2365–2373. (doi:10.4049/jimmunol.1500542) [DOI] [PubMed] [Google Scholar]
- 78.Nastase MV, et al. 2016. An essential role for SHARPIN in the regulation of caspase 1 activity in sepsis. Am. J. Pathol. 186, 1206–1220. (doi:10.1016/j.ajpath.2015.12.026) [DOI] [PubMed] [Google Scholar]
- 79.Shimizu S, Fujita H, Sasaki Y, Tsuruyama T, Fukuda K, Iwai K. 2016. Differential involvement of the Npl4 zinc finger domains of SHARPIN and HOIL-1L in linear ubiquitin chain assembly complex-mediated cell death protection. Mol. Cell. Biol. 36, 1569–1583. (doi:10.1128/MCB.01049-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zinngrebe J, et al. 2016. LUBAC deficiency perturbs TLR3 signaling to cause immunodeficiency and autoinflammation. J. Exp. Med. 213, 2671–2689. (doi:10.1084/jem.20160041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Damgaard RB, et al. 2013. Disease-causing mutations in the XIAP BIR2 domain impair NOD2-dependent immune signalling. EMBO Mol. Med. 5, 1278–1295. (doi:10.1002/emmm.201303090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Damgaard RB, et al. 2012. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol. Cell. 46, 746–758. (doi:10.1016/j.molcel.2012.04.014) [DOI] [PubMed] [Google Scholar]
- 83.Emmerich CH, Bakshi S, Kelsall IR, Ortiz-Guerrero J, Shpiro N, Cohen P. 2016. Lys63/Met1-hybrid ubiquitin chains are commonly formed during the activation of innate immune signalling. Biochem. Biophys. Res. Commun. 474, 452–461. (doi:10.1016/j.bbrc.2016.04.141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zak DE, et al. 2011. Systems analysis identifies an essential role for SHANK-associated RH domain-interacting protein (SHARPIN) in macrophage Toll-like receptor 2 (TLR2) responses. Proc. Natl Acad. Sci. USA 108, 11 536–11 541. (doi:10.1073/pnas.1107577108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rodgers MA, et al. 2014. The linear ubiquitin assembly complex (LUBAC) is essential for NLRP3 inflammasome activation. J. Exp. Med. 211, 1333–1347. (doi:10.1084/jem.20132486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bowman J, Rodgers MA, Shi M, Amatya R, Hostager B, Iwai K, Gao SJ, Jung JU. 2015. Posttranslational modification of HOIP blocks Toll-like receptor 4-mediated linear-ubiquitin-chain formation. mBio 6, e01777-15. (doi:10.1128/mBio.01777-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Belgnaoui SM, et al. 2012. Linear ubiquitination of NEMO negatively regulates the interferon antiviral response through disruption of the MAVS-TRAF3 complex. Cell Host Microbe 12, 211–222. (doi:10.1016/j.chom.2012.06.009) [DOI] [PubMed] [Google Scholar]
- 88.Chattopadhyay S, Kuzmanovic T, Zhang Y, Wetzel JL, Sen GC. 2016. Ubiquitination of the transcription factor IRF-3 activates RIPA, the apoptotic pathway that protects mice from viral pathogenesis. Immunity 44, 1151–1161. (doi:10.1016/j.immuni.2016.04.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang L, Wang Y, Zhao J, Ren J, Hall KH, Moorman JP, Yao ZQ, Ning S. 2016. LUBAC modulates LMP1 activation of NF-κB and IRF7. J. Virol. 91, e01138-16. (doi:10.1128/JVI.01138-16) [Google Scholar]
- 90.Inn KS, Gack MU, Tokunaga F, Shi M, Wong LY, Iwai K, Jung JU. 2011. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol. Cell. 41, 354–365. (doi:10.1016/j.molcel.2010.12.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klein T, et al. 2015. The paracaspase MALT1 cleaves HOIL1 reducing linear ubiquitination by LUBAC to dampen lymphocyte NF-κB signalling. Nat. Commun. 6, 8777 (doi:10.1038/ncomms9777) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Elton L, Carpentier I, Staal J, Driege Y, Haegman M, Beyaert R. 2016. MALT1 cleaves the E3 ubiquitin ligase HOIL-1 in activated T cells, generating a dominant negative inhibitor of LUBAC-induced NF-κB signaling. FEBS J. 283, 403–412. (doi:10.1111/febs.13597) [DOI] [PubMed] [Google Scholar]
- 93.Hailfinger S, Schmitt A, Schulze-Osthoff K. 2016. The paracaspase MALT1 dampens NF-κB signalling by cleaving the LUBAC subunit HOIL-1. FEBS J. 283, 400–402. (doi:10.1111/febs.13639) [DOI] [PubMed] [Google Scholar]
- 94.Douanne T, Gavard J, Bidere N. 2016. The paracaspase MALT1 cleaves the LUBAC subunit HOIL1 during antigen receptor signaling. J. Cell Sci. 129, 1775–1780. (doi:10.1242/jcs.185025) [DOI] [PubMed] [Google Scholar]
- 95.Yang YK, Yang C, Chan W, Wang Z, Deibel KE, Pomerantz JL. 2016. Molecular determinants of scaffold-induced linear ubiquitinylation of B cell lymphoma/leukemia 10 (Bcl10) during T cell receptor and oncogenic caspase recruitment domain-containing protein 11 (CARD11) signaling. J. Biol. Chem. 291, 25 921–25 936. (doi:10.1074/jbc.M116.754028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Teh CE, et al. 2016. Linear ubiquitin chain assembly complex coordinates late thymic T-cell differentiation and regulatory T-cell homeostasis. Nat. Commun. 7, 13353 (doi:10.1038/ncomms13353) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sasaki Y, et al. 2013. Defective immune responses in mice lacking LUBAC-mediated linear ubiquitination in B cells. EMBO J. 32, 2463–2476. (doi:10.1038/emboj.2013.184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Niu J, Shi Y, Iwai K, Wu ZH. 2011. LUBAC regulates NF-κB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J. 30, 3741–3753. (doi:10.1038/emboj.2011.264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Asaoka T, et al. 2016. Linear ubiquitination by LUBEL has a role in Drosophila heat stress response. EMBO Rep. 17, 1624–1640. (doi:10.15252/embr.201642378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Minakhina S, Yang J, Steward R. 2003. Tamo selectively modulates nuclear import in Drosophila. Genes Cells 8, 299–310. (doi:10.1046/j.1365-2443.2002.00634.x) [DOI] [PubMed] [Google Scholar]
- 101.Rantala JK, et al. 2011. SHARPIN is an endogenous inhibitor of beta1-integrin activation. Nat. Cell Biol. 13, 1315–1324. (doi:10.1038/ncb2340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.De Franceschi N, Peuhu E, Parsons M, Rissanen S, Vattulainen I, Salmi M, Ivaska J, Pouwels J. 2015. Mutually exclusive roles of SHARPIN in integrin inactivation and NF-κB signaling. PLoS ONE 10, e0143423 (doi:10.1371/journal.pone.0143423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Peuhu E, et al. 2017. SHARPIN regulates collagen architecture and ductal outgrowth in the developing mouse mammary gland. EMBO J. 36, 165–182. (doi:10.15252/embj.201694387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boisson B, et al. 2015. Human HOIP and LUBAC deficiency underlies autoinflammation, immunodeficiency, amylopectinosis, and lymphangiectasia. J. Exp. Med. 212, 939–951. (doi:10.1084/jem.20141130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nilsson J, et al. 2013. Polyglucosan body myopathy caused by defective ubiquitin ligase RBCK1. Ann. Neurol. 74, 914–919. (doi:10.1002/ana.23963) [DOI] [PubMed] [Google Scholar]
- 106.Wang K, et al. 2013. Whole-genome DNA/RNA sequencing identifies truncating mutations in RBCK1 in a novel Mendelian disease with neuromuscular and cardiac involvement. Genome Med. 5, 67 (doi:10.1186/gm471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou Q, et al. 2016. Biallelic hypomorphic mutations in a linear deubiquitinase define otulipenia, an early-onset autoinflammatory disease. Proc. Natl Acad. Sci. USA 113, 10 127–10 132. (doi:10.1073/pnas.1612594113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang Y, et al. 2016. Targeting non-proteolytic protein ubiquitination for the treatment of diffuse large B cell lymphoma. Cancer Cell. 29, 494–507. (doi:10.1016/j.ccell.2016.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang Y, et al. 2014. Essential role of the linear ubiquitin chain assembly complex in lymphoma revealed by rare germline polymorphisms. Cancer Discov. 4, 480–493. (doi:10.1158/2159-8290.CD-13-0915) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lewis M, Vyse S, Shields A, Boeltz S, Gordon P, Spector T, Lehner P, Walczak H, Vyse T. 2015. Effect of UBE2L3 genotype on regulation of the linear ubiquitin chain assembly complex in systemic lupus erythematosus. Lancet 385, S9 (doi:10.1016/S0140-6736(15)60324-5) [DOI] [PubMed] [Google Scholar]
- 111.Lewis MJ, Vyse S, Shields AM, Boeltz S, Gordon PA, Spector TD, Lehner PJ, Walczak H, Vyse TJ. 2015. UBE2L3 polymorphism amplifies NF-κB activation and promotes plasma cell development, linking linear ubiquitination to multiple autoimmune diseases. Am. J. Hum. Genet. 96, 221–234. (doi:10.1016/j.ajhg.2014.12.024) [DOI] [PMC free article] [PubMed] [Google Scholar]