Figure 2. Principal component and haplotype analysis of families harboring c.424G>A p.Val142Ile variant and schematic illustrating the newly identified and previously reported RNASEH1 mutations to date.
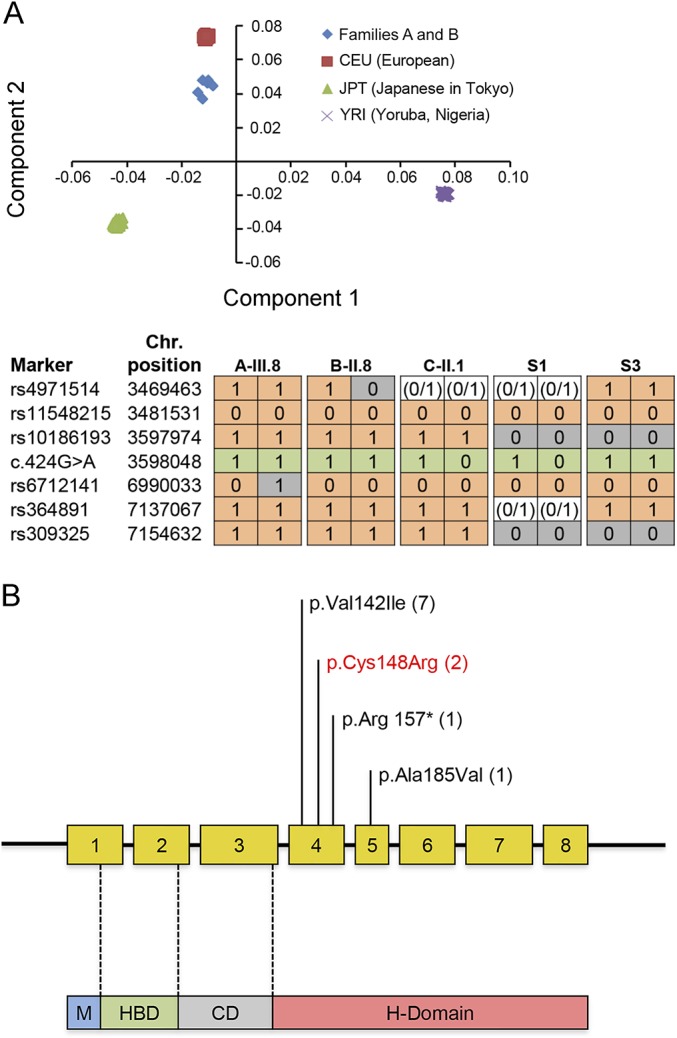
(A) Principal component analysis (top panel). X-axis represents component 1; Y-axis represents component 2. Families A and B cluster to the same ethnic group. Haplotype analysis of individuals harboring the RNASEH1 mutation c.424G>A p.Val142Ile (bottom panel). “1” indicates the presence of the marker, “0” indicates the absence of the marker, and “0/1” is used when haplotype could not be phased. Haplotypes reported are for A-III.8, B-II.8, C-II.1, S-1, and S-3. Green = c.424G>A mutation; gray = markers differing from reference haplotype. Families A and B shared a haplotype of at least 3.57 Mb. However, 2 additional haplotypes were different, suggesting distant recombination events or that the haplotypes arose independently. (B) The RNASEH1 gene has 8 exons encoding 4 protein domains as follows: mitochondrial targeting sequencing domain (M), hybrid-binding domain (HBD), connection domain (CD), and ribonuclease H domain (H-domain). The latter conducts all catalytic activity. Both herein newly identified and previously reported mutations are illustrated in the schematic. To date, all affected individuals harbor the “common” c.424G>A p.Val142Ile mutation either in homozygous or heterozygous states. Brackets = number of families with each mutation. Red = newly reported variant.
